













| General Information: |
|
| Name(s) found: |
NTHL1_MOUSE
[Swiss-Prot]
NTH1_MOUSE [Swiss-Prot] |
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Mus musculus |
| Length: | 300 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
mitochondrion [IDA] |
| Biological Process: |
DNA repair
[TAS]
base-excision repair [IEA] nucleotide-excision repair, DNA incision, 5'-to lesion [IDA] metabolic process [IEA] response to DNA damage stimulus [IEA] |
| Molecular Function: |
DNA binding
[IEA]
metal ion binding [IEA] catalytic activity [IEA] endonuclease activity [IEA] lyase activity [IEA] DNA-(apurinic or apyrimidinic site) lyase activity [IDA] DNA N-glycosylase activity [IDA] 4 iron, 4 sulfur cluster binding [IEA] iron-sulfur cluster binding [IEA] hydrolase activity [IEA] hydrolase activity, acting on glycosyl bonds [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSGVRMVTR SRSRATRIAS EGCREELAPR EAAAEGRKSH RPVRHPRRTQ KTHVAYEAAN 60
61 GEEGEDAEPL KVPVWEPQNW QQQLANIRIM RSKKDAPVDQ LGAEHCYDAS APPKVRRYQV 120
121 LLSLMLSSQT KDQVTAGAMQ RLRARGLTVE SILQTDDDTL GRLIYPVGFW RNKVKYIKQT 180
181 TAILQQRYEG DIPASVAELV ALPGVGPKMA HLAMAVAWGT ISGIAVDTHV HRIANRLRWT 240
241 KKMTKTPEET RKNLEEWLPR VLWSEVNGLL VGFGQQICLP VHPRCQACLN KALCPAAQDL 300
301 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
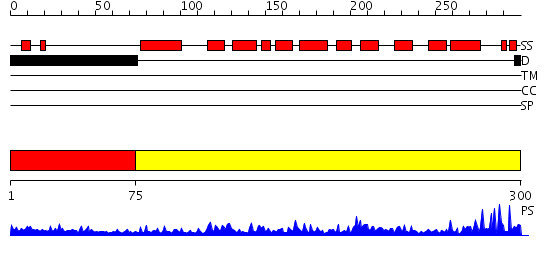
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [75..300] | 58.09691 | REFINEMENT OF THE NATIVE STRUCTURE OF ENDONUCLEASE III TO A RESOLUTION OF 1.85 ANGSTROM |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)