













| General Information: |
|
| Name(s) found: |
HSF_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 609 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIQTASTISS NGGTNQGMES SSANSPEMNG TQNSMSVGMS GSGSSQNRKI TQFSNKLYNM 60
61 VNDSSTDSLI RWSDRGDSFL VIGHEDFAKL VLPRYFKHNN FSSFVRQLNM YGFHKVPHIQ 120
121 QGVLQSDSPN ELLEFANPNF QRDQPELLCL VTRKKAGSQP VEESNTSLDM STISSELQNI 180
181 RIQQMNLSNE LSRIQVDNAA LWQENMENRE RQRRHQETID KILRFLASVY LDGKQKPPSK 240
241 VMPKSRRLLL EAKYPTVSPT NEPSAHSRPS PQGTTANSSS ASISSLHNTT PDGEGKYRSV 300
301 QNGRALNYVS SFNSDSHSPK DYISQSYTNE PGLKKESADS FNNSIDSYIS PNQSPNTDVP 360
361 SLNRDDTTDP KVVNTGDIIN MLDDANSIEG SNMNSLSPLL FDYPNSLYPV NNTSSEQHHN 420
421 SYRGSVSNSQ PSGNLSESTN LQPVQPVDYM SNSNPSYGSY NAEDQLTNFH PGYAMDQKRI 480
481 SKLSDGITKQ DQNIQALADI LGIPLGDGKI DDAGFSANSP TNLNLPVSSD LDSVLNIPPN 540
541 EDVFPDSNPV FDEFTNISNL TSPIEASNGN TFGSNPVSLP NQQKSVNPSL MTVSSPRQVR 600
601 KKRKSSIGA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
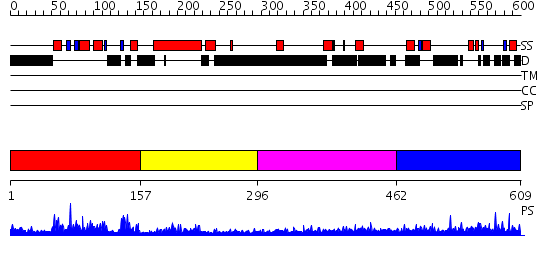
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | 39.0 | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | |
| 2 | View Details | [157..295] | 9.167491 | No description for PF00447.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [296..461] | 1.154994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [462..609] | 2.053994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)