













| General Information: |
|
| Name(s) found: |
DHX15_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 735 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPAQKKLRQ ESKNPYLAHL NNGDDSEEVV SSKGLTRRAT TVAQAAKAEE GPNNFFNDKP 60
61 FSQNYFKILE TRRELPVYQQ REEFLKIYHE NQIIVFVGET GSGKTTQIPQ FVLYDELPHL 120
121 TNTQIACTQP RRVAAMSVAK RVADEMDVDL GEEVGYNIRF EDCSGPNTLL KYMTDGMLLR 180
181 EAMTDHMLSR YSCIILDEAH ERTLATDILM GLMKRLATRR PDLKIIVMSA TLDAKKFQKY 240
241 FFDAPLLAVP GRTYPVEIYY TQEPERDYLE AALRTVLQIH VEEGPGDILV FLTGEEEIED 300
301 ACRKITLEAD DLVREGAAGP LKVYPLYGSL PPNQQQRIFE PTPEDTKSGY GRKVVISTNI 360
361 AETSLTIDGI VYVVDPGFSK QKIYNPRIRV ESLLVSPISK ASAQQRAGRA GRTRPGKCFR 420
421 LYTEEAFRKE LIEQTYPEIL RSNLSSTVLE LKKLGIDDLV HFDYMDPPAP ETMMRALEEL 480
481 NYLNCLDDNG DLTPLGRKAS EFPLDPNLAV MLIRSPEFYC SNEVLSLTAL LSVPNVFVRP 540
541 NSARKLADEM RQQFTHPDGD HLTLLNVYHA YKSGEGTADW CWNHFLSHRA LISADNVRKQ 600
601 LRRTMERQEV ELISTPFDDK NYYVNIRRAL VSGFFMQVAK KSANGKNYVT MKDNQVVSLH 660
661 PSCGLSVTPE WVVYNEFVLT TKSFIRNVTA IRPEWLIELA PNYYDLDDFD NNKEVKSALQ 720
721 KVYQMAARSK KNARR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
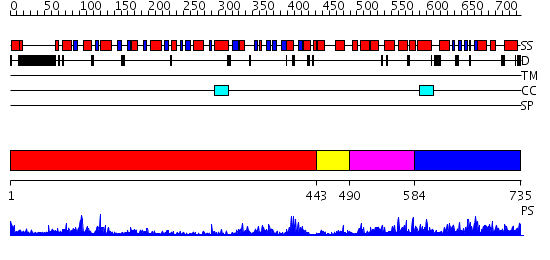
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..442] | 61.69897 | No description for 2j0qA was found. | |
| 2 | View Details | [443..489] | 10.30103 | No description for 2d7dA was found. | |
| 3 | View Details | [490..583] | 1.13 | No description for 2v8oA was found. | |
| 4 | View Details | [584..735] | 29.886057 | No description for PF07717.7 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)