













| General Information: |
|
| Name(s) found: |
FHL1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 743 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPVAEIKNAT QQPSSTNRVQ AYAKLEFEKF SFFVQTLQVT MGRKASNSSD CDVHLGDTKA 60
61 ISRQHAKIFY SFPNQRFEIS VMGKNGAFVD GEFVERGKSV PLRSGTRVQI GQISFSFLLP 120
121 EGSEEDGHLK ETGITPLSLQ QGKIAYSDEF GGKPTGSFHT VTSNQEKDLL FSHIKHESDL 180
181 PLGLSPADTN ISNATSIIEH PDAANAHTLA SLNQPPKHLT VSPSSIQRLS PQPYVRPTSD 240
241 ERPIETDSSV SAPKVANHDE ELKQGKSTSP SDTVLHPDLN GSPDTGDATQ KPNLSYANLI 300
301 ARTLIANPNK KMTLGDICEW IANNWSYYRH QPPAWHNSIR HNLSLNKAFI RIPRRQNEPG 360
361 KGSFWMLDPS YIDQFEGNFF RRTKKPTPSA TPAAHPDTAR ENELAAIQTK GISAGKTEQL 420
421 NPQKETSRSK THTSRGENVE DRPQSLLQNG IQPIIMRDGK LALNPEFFKN ANGEQQAPNE 480
481 QAVQAISLLQ QHINKQLGPA AANNPEQATA IANALAVALA QKLQKQQTQM QGPQQVQQQA 540
541 KRRKAYTSQQ LNPAPTAMPH PNITSPSPSI SVTQRPAVNV GPPPYVRPSA PSKLPDTRQS 600
601 IGDPLPPGAM ANVSAGPSSV RSSSYNSTAS ESKSEITSHQ NLHTIPINKP FTSDRPLYSS 660
661 PNDTLERVET GNQGQRMNSI GNASSFSKRD IMENENGSFD TNAKNGNNVD DSSSVRGMNL 720
721 PSNSSDALRG VKRPLDETSS SYT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
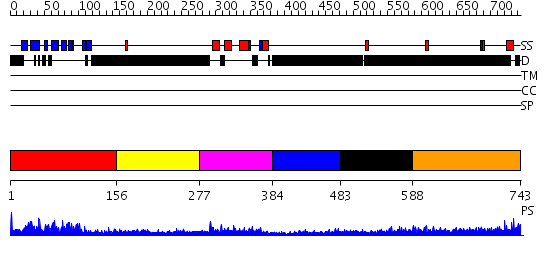
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] | 13.39794 | Cell cycle checkpoint protein Chfr | |
| 2 | View Details | [156..276] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [277..383] | 31.045757 | Adipocyte-transcription factor FREAC-11 (s12, fkh-14) | |
| 4 | View Details | [384..482] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [483..587] | 3.303997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [588..743] | 4.229988 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)