













| General Information: |
|
| Name(s) found: |
ATH1 /
YPR026W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1211 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type cell wall
[IDA cell wall-bounded periplasmic space [IDA fungal-type vacuole [IMP |
| Biological Process: |
trehalose catabolic process
[IDA response to stress [IMP |
| Molecular Function: |
alpha,alpha-trehalase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRIRSLWFN AEASYSNLNN SPSLRNKNST GNNSRSKNYR SFSRFDLINS ILLLMMLFLL 60
61 AIFVTALYLT KSSRLTYSHA SRAALFNPLG VISPSLGNHT LNYDPEARES SKKLYELLSD 120
121 FNTAYYDDEN MILGSNLFSK NTYSRQPYVA NGYIGSRIPN IGFGYALDTL NFYTDAPGAL 180
181 NNGWPLRNHR FAGAFVSDFY CLQPKLNSTN FPELDDVGYS TVISSIPQWT NLQFSLVNDS 240
241 KWFNPQNVTL DDVTNYSQNL SMKDGIVTTE LDWLNSQIHV KSEIWAHRHI HPLGVVSLEI 300
301 SLNTDHLPSD FDSLDVNIWD ILDFNTSHRT VLHSTGTDEK NNAVFMIVQP DNVPSSNCAI 360
361 YSTCTVKYEN STNPINSSES FEEKDVSSNI YNVILTEDQP KIIVHKYVGI MSTEFNKNKE 420
421 QQDNTNIGLA KMIALNSKGN YEKLLSSHKR AWYDLYNDAF IEIPSDSLLE MTARSSLFHL 480
481 LANTRDYNVS SDRGLPVGVS GLSSDSYGGM VFWDADIWME PALLPFFPNV AQNMNNYRNA 540
541 THSQAKLNAE KYGYPGAIYP WTSGKYANCT STGPCVDYEY HINVDVAMAS FSIYLNGHEG 600
601 IDDEYLRYTT WPIIKNAAQF FTAYVKYNSS LGLYETYNLT DPDEFANHIN NGAFTNAGIK 660
661 TLLKWATDIG NHLGEVVDPK WSEISKDIYI PRSSSNITLE YSGMNSSVEI KQADVTLMVY 720
721 PLGYINDESI LNNAIKDLYY YSERQSASGP AMTYPVFVAA AAGLLNHGSS SQSYLYKSVL 780
781 PYLRAPFAQF SEQSDDNFLT NGLTQPAFPF LTANGGFLQS ILFGLTGIRY SYEVDPDTKK 840
841 INRLLRFNPI ELPLLPGGIA IRNFKYMNQV LDIIIDDHNG TIVHKSGDVP IHIKIPNRSL 900
901 IHDQDINFYN GSENERKPNL ERRDVDRVGD PMRMDRYGTY YLLKPKQELT VQLFKPGLNA 960
961 RNNIAENKQI TNLTAGVPGD VAFSALDGNN YTHWQPLDKI HRAKLLIDLG EYNEKEITKG1020
1021 MILWGQRPAK NISISILPHS EKVENLFANV TEIMQNSGND QLLNETIGQL LDNAGIPVEN1080
1081 VIDFDGIEQE DDESLDDVQA LLHWKKEDLA KLIEQIPRLN FLKRKFVKIL DNVPVSPSEP1140
1141 YYEASRNQSL IEILPSNRTT FTIDYDKLQV GDKGNTDWRK TRYIVVAVQG VYDDYDDDNK1200
1201 GATIKEIVLN D |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
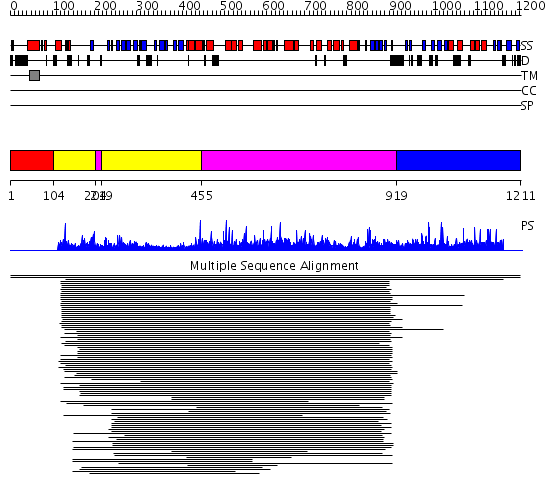
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [104..203] [219..454] |
1186.9897 | Lactobacillus maltose phosphorylase, central domain; Lactobacillus maltose phosphorylase, N-terminal domain | |
| 3 | View Details | [204..218] [455..918] |
1186.9897 | Lactobacillus maltose phosphorylase, central domain; Lactobacillus maltose phosphorylase, N-terminal domain | |
| 4 | View Details | [919..1211] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|