













| General Information: |
|
| Name(s) found: |
VTS1 /
YOR359W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 523 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
regulation of mRNA stability
[IMP protein targeting to vacuole [IGI |
| Molecular Function: |
RNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKHPYEEFPT GSKSPYNMSR GAHPGAVLLS PQSSAINKNN PGSNSGNNQG NSSVTANVLS 60
61 PQSHSMSLND MLDQQSFMLD TAGTRAQPLQ QQQQQQQQQQ QASLPSLNIQ TVSSTAAGSA 120
121 IVSPMMQSPK ALQSTLSSTS MYLDSFQRSP NNILGIPSQS GSIPLPQSRQ SQQQSQSQKN 180
181 DPNMGTNFSQ DINQLCSWIS MLNSSQQNTV MDNILSILND DVLKYTKLKI ETLTNTPFIS 240
241 PPLPAIASPI PNRDDTQILN IDSVFSSSPI TNDPENTDNL LYQNWSPQPH SIPISQPIYD 300
301 NITDASQRSK SAEPHVNSSP NLIPVQKQFN NGNSTKYKKL PSENPNYLSH SLSSSHSFFQ 360
361 PKKRSNMGNE YNSHHHHSLH HPLHNTTSYF SNTSRPSGTD LNKSNQNVFN NTITHPNAGP 420
421 TSATSTSTSS NGNTPLSSNS SMNPKSLTDP KLLKNIPMWL KSLRLHKYSD ALSGTPWIEL 480
481 IYLDDETLEK KGVLALGARR KLLKAFGIVI DYKERDLIDR SAY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ASE1 RPS25B STM1 VTS1 YPL150W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
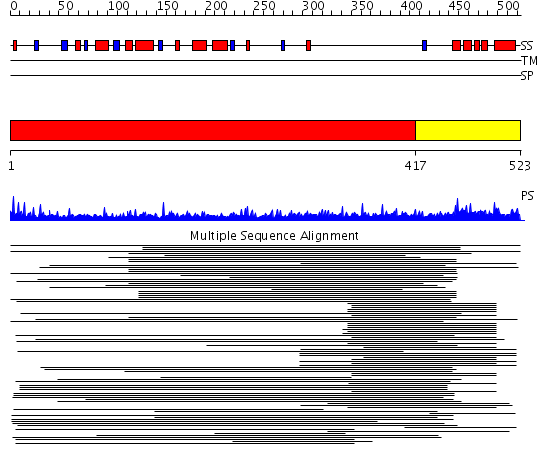
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..416] | 21.281997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [417..523] | 14.158997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [309..407] | 2.14599 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [408..523] | 28.0 | RNA recognition by the Vts1 SAM domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)