













| General Information: |
|
| Name(s) found: |
PDR17 /
YNL264C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 350 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
response to drug
[IGI phospholipid biosynthetic process [IMP phospholipid transport [IDA |
| Molecular Function: |
phosphatidylinositol transporter activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLFSRKRDH TPAVPKEKLI PCDKIFLDPP AKYGNAPLLE PISEDQNEKY RAVLRHFQDD 60
61 DLKLPENLND LDNGTHANDR PLSDWEKFWL SRECFLRYLR ANKWNTANAI KGLTKTLVWR 120
121 REIGLTHGKE DKDPLTADKV AVENETGKQV ILGFDNAKRP LYYMKNGRQN TESSFRQVQE 180
181 LVYMMETATT VAPQGVEKIT VLVDFKSYKE PGIITDKAPP ISIARMCLNV MQDHYPERLA 240
241 KCVLINIPWF AWAFLKMMYP FLDPATKAKA IFDEPFENHI EPSQLDALYN GLLDFKYKHE 300
301 VYWPDMVKKV DDLRLKRFDR FLKFGGIVGL SEYDTKGQHD ELKYPVDMVI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
PDR17 PSD2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
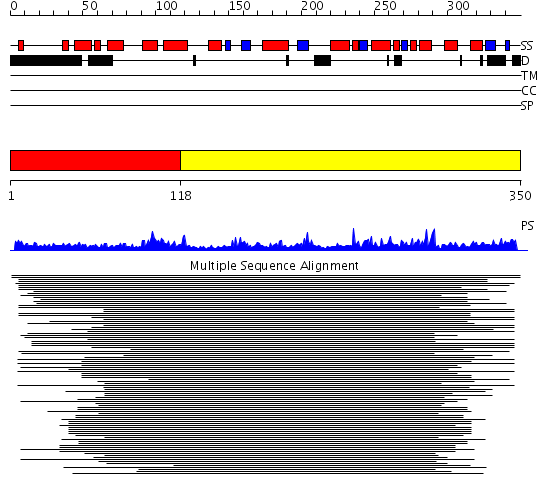
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 113.68867 | N-terminal domain of phosphatidylinositol transfer protein sec14p; C-terminal domain of phosphatidylinositol transfer protein sec14p | |
| 2 | View Details | [118..350] | 113.68867 | N-terminal domain of phosphatidylinositol transfer protein sec14p; C-terminal domain of phosphatidylinositol transfer protein sec14p |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)