













| General Information: |
|
| Name(s) found: |
MSS11 /
YMR164C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
pseudohyphal growth
[IGI starch catabolic process [IMP positive regulation of transcription from RNA polymerase II promoter [IDA |
| Molecular Function: |
specific RNA polymerase II transcription factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNTTNINTN ERSSNTDFSS APNIKGLNSH TQLQFDADSR VFVSDVMAKN SKQLLYAHIY 60
61 NYLIKNNYWN SAAKFLSEAD LPLSRINGSA SGGKTSLNAS LKQGLMDIAS KGDIVSEDGL 120
121 LPSKMLMDAN DTFLLEWWEI FQSLFNGDLE SGYQQDHNPL RERIIPILPA NSKSNMPSHF 180
181 SNLPPNVIPP TQNSFPVSEE SFRPNGDGSN FNLNDPTNRN VSERFLSRTS GVYDKQNSAN 240
241 FAPDTAINSD IAGQQYATIN LHKHFNDLQS PAQPQQSSQQ QIQQPQHQPQ HQPQQQQQQQ 300
301 QQQQQQQQQQ QQQQQQQQQQ QQQHQQQQQT PYPIVNPQMV PHIPSENSHS TGLMPSVPPT 360
361 NQQFNAQTQS SMFSDQQRFF QYQLHHQNQG QAPSFQQSQS GRFDDMNAMK MFFQQQALQQ 420
421 NSLQQNLGNQ NYQSNTRNNT AEETTPTNDN NANGNSLLQE HIRARFNKMK TIPQQMKNQS 480
481 TVANPVVSDI TSQQQYMHMM MQRMAANQQL QNSAFPPDTN RIAPANNTMP LQPGNMGSPV 540
541 IENPGMRQTN PSGQNPMINM QPLYQNVSSA MHAFAPQQQF HLPQHYKTNT SVPQNDSTSV 600
601 FPLPNNNNNN NNNNNNNNNN NSNNSNNNNN NNNNNNNSNN TPTVSQPSSK CTSSSSTTPN 660
661 ITTTIQPKRK QRVGKTKTKE SRKVAAAQKV MKSKKLEQNG DSAATNFINV TPKDSGGKGT 720
721 VKVQNSNSQQ QLNGSFSMDT ETFDIFNIGD FSPDLMDS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #27 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps3-5 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
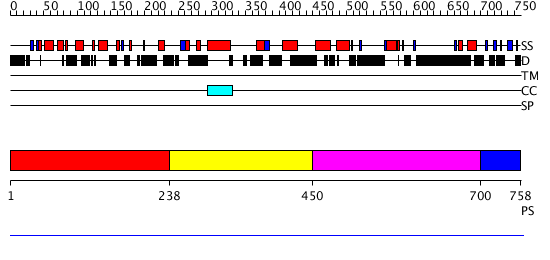
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..237] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [238..449] | 1.002995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [450..699] | 2.004996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [700..758] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)