













| General Information: |
|
| Name(s) found: |
YKL171W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 928 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
proteolysis
[IPI |
| Molecular Function: |
protein kinase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFTSQRQLRQ NGSPMSSSRS SQHSSGTASP ISDSPASNRS YGRDLRGLMG IDIPANEPAF 60
61 NRANSSDTIY FRPKKIYKME HEHPSRSTLV QLQTRSQPDD VASSQVNPEG GTDDLELGDP 120
121 CGNQSLYTIG AEYVPDLDFT KLVNEWQKST EDLYEFRSSA TPQVQIKDSG KGNYELWSSP 180
181 DAILTQNKLR RDSFSQENSD SLSPEDSILS RNLHSKVKPI PLPRNSQQIF TPLSNLEAER 240
241 RSSYTTSSNN NSITQNNKFS FAKLKYSLPT QSSAVPASFD SNASSLNFLP TTTLSTLSEL 300
301 QISPNDMMDL IQKLPRNFLN LPYTQRKKVI IEHAPSHDYK AMMSLVKKFM LTSSRSNFSL 360
361 AGFANNASVS QATANDDNIN SRNTPNNSND TYVNTRPLQR SRHGSIASQF LSSFSPSMTS 420
421 IAKMNSNPLS GSAGGSARPD DKGMEILGHR LGKIIGFGAW GIIRECFDIE TGVGRVIKIV 480
481 KFKGHQNIKK HVLREVAIWR TLKHNRILPL LDWKLDDNYA MYCLTERIND GTLYDLVISW 540
541 DEFKRSKIPF AERCRLTIFL SLQLLSALKY MHSKTIVHGD IKLENCLLQK EGKKSDWKVF 600
601 LCDFGMSCHF DEKHVYRNDT FDENLSSGNS HRKRKSIEQT NLIKYPTTNF LPDDRTNDFD 660
661 ASENLKYQFE NRKHQPFTPK GMVSSSSHSL KHLNQPSSSS SSNLFHKPAS QPQPQHRSPF 720
721 HGRHKTTDFS NLEPEPSKYI GSLPYASPEL LRYSDARRSK SVEMHIYDSP DSSQSEISAA 780
781 SSSSSNLSSL SSSTKASAVT NSGVTTSSPS GSSTDFPCIV SPLGPASDIW ALGVMLYTML 840
841 VGKLPFNHEF EPRLRSLIKV GEFDRFSLAQ VCKFDRKKNE GTIGQGLYDT VIGCLTIDLD 900
901 KRWKLKRIEE VLQNEMNLSE AIHDNNGS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
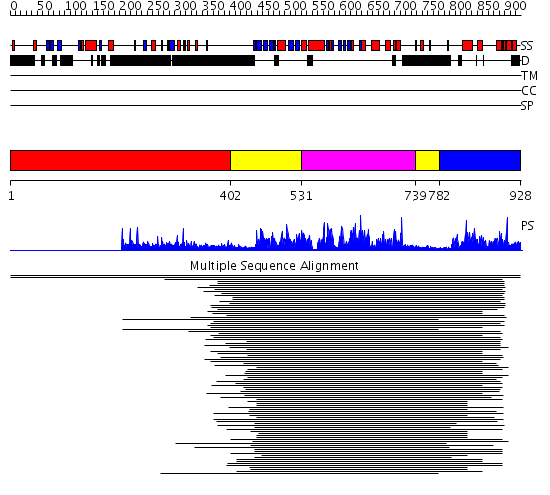
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..401] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [402..530] [739..781] |
167.54902 | Calmodulin-dependent protein kinase | |
| 3 | View Details | [531..738] | 167.54902 | Calmodulin-dependent protein kinase | |
| 4 | View Details | [782..928] | 1.326968 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [780..928] | 7.41 | Crystal structure of the protein kinase domain of yeast AMP-activated protein kinase Snf1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)