













| General Information: |
|
| Name(s) found: |
PGM1 /
YKL127W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 570 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[TAS]
|
| Biological Process: |
carbohydrate metabolic process
[IEA]
glycogen biosynthetic process [IGI] glucose 6-phosphate metabolic process [ISS] galactose catabolic process [IGI] glucose 1-phosphate metabolic process [IGI][ISS] trehalose biosynthetic process [IGI] UDP-glucose metabolic process [IGI] glucose metabolic process [IEA] |
| Molecular Function: |
phosphoglucomutase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLLIDSVPT VAYKDQKPGT SGLRKKTKVF MDEPHYTENF IQATMQSIPN GSEGTTLVVG 60
61 GDGRFYNDVI MNKIAAVGAA NGVRKLVIGQ GGLLSTPAAS HIIRTYEEKC TGGGIILTAS 120
121 HNPGGPENDL GIKYNLPNGG PAPESVTNAI WEASKKLTHY KIIKNFPKLN LNKLGKNQKY 180
181 GPLLVDIIDP AKAYVQFLKE IFDFDLIKSF LAKQRKDKGW KLLFDSLNGI TGPYGKAIFV 240
241 DEFGLPAEEV LQNWHPLPDF GGLHPDPNLT YARTLVDRVD REKIAFGAAS DGDGDRNMIY 300
301 GYGPAFVSPG DSVAIIAEYA PEIPYFAKQG IYGLARSFPT SSAIDRVAAK KGLRCYEVPT 360
361 GWKFFCALFD AKKLSICGEE SFGTGSNHIR EKDGLWAIIA WLNILAIYHR RNPEKEASIK 420
421 TIQDEFWNEY GRTFFTRYDY EHIECEQAEK VVALLSEFVS RPNVCGSHFP ADESLTVIDC 480
481 GDFSYRDLDG SISENQGLFV KFSNGTKFVL RLSGTGSSGA TIRLYVEKYT DKKENYGQTA 540
541 DVFLKPVINS IVKFLRFKEI LGTDEPTVRT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
AHA1 HOM3 NTG1 OPY1 PGM1 PGM2 PRB1 RAD59 RPB3 RPT3 SEC27 |
| View Details | Qiu et al. (2008) |

|
PGM1 PGM2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
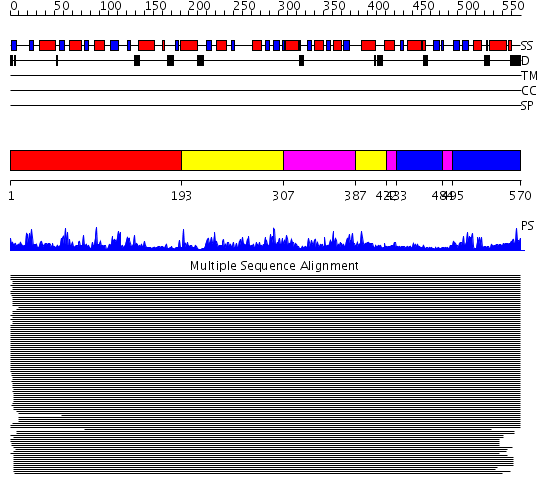
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..192] | 1672.0 | Phosphoglucomutase | |
| 2 | View Details | [193..306] [387..421] |
1672.0 | Phosphoglucomutase | |
| 3 | View Details | [307..386] [422..432] [484..494] |
1672.0 | Phosphoglucomutase | |
| 4 | View Details | [433..483] [495..570] |
1672.0 | Phosphoglucomutase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)