













| General Information: |
|
| Name(s) found: |
AAT1 /
YKL106W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 451 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
asparagine biosynthetic process from oxaloacetate
[IC]
aspartate biosynthetic process [IC] aspartate catabolic process [TAS] |
| Molecular Function: |
L-aspartate:2-oxoglutarate aminotransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRTRLTNCS LWRPYYTSSL SRVPRAPPDK VLGLSEHFKK VKNVNKIDLT VGIYKDGWGK 60
61 VTTFPSVAKA QKLIESHLEL NKNLSYLPIT GSKEFQENVM KFLFKESCPQ FGPFYLAHDR 120
121 ISFVQTLSGT GALAVAAKFL ALFISRDIWI PDPSWANHKN IFQNNGFENI YRYSYYKDGQ 180
181 IDIDGWIEQL KTFAYNNQQE NNKNPPCIIL HACCHNPTGL DPTKEQWEKI IDTIYELKMV 240
241 PIVDMAYQGL ESGNLLKDAY LLRLCLNVNK YPNWSNGIFL CQSFAKNMGL YGERVGSLSV 300
301 ITPATANNGK FNPLQQKNSL QQNIDSQLKK IVRGMYSSPP GYGSRVVNVV LSDFKLKQQW 360
361 FKDVDFMVQR LHHVRQEMFD RLGWPDLVNF AQQHGMFYYT RFSPKQVEIL RNNYFVYLTG 420
421 DGRLSLSGVN DSNVDYLCES LEAVSKMDKL A |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
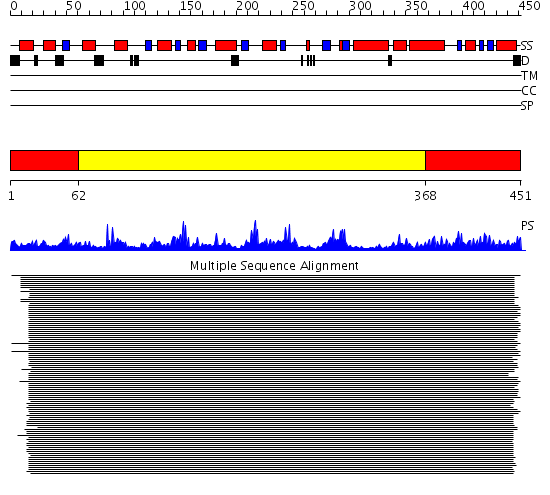
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] [368..451] |
817.927757 | Aspartate aminotransferase, AAT | |
| 2 | View Details | [62..367] | 817.927757 | Aspartate aminotransferase, AAT |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)