













| General Information: |
|
| Name(s) found: |
GSM1 /
YJL103C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 618 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
oxidative phosphorylation
[IEP |
| Molecular Function: |
DNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKKLPSELK QTRKSIQTAC EFCHTKHIQC DVGRPCQNCL KRNIGKFCRD KKRKSRKRIE 60
61 KHGTQPYLNL GKRLVIHDVP SKTVSPSSVH LQRDFLSSDQ EKPGKTPAHN TNIQYTYNIN 120
121 DNFQSAGSIP RITNFNTNNR QTVLENTSNN ISASQAVHLM NDPIIPTVRK STLNLKSHFL 180
181 EQHKAMQQPL ATNCLVATSN VPVHSGMDDS NKSDDDVDDE TNIHFDSMWC NDEYMKLKDI 240
241 VDISTPFLPN NSQIFSLQES EYPNPSASTR GNSSLHLTNL LNSTKSVNDQ KDSSIGHSTS 300
301 TFNTYDEVVS RPFISLDMLH LNRGANANTH PSHNAKLESE CDSSSHSDAD LEKHDTDFIS 360
361 PSKFRELVKT PQDLYDNKCL IKPHNYKLAY TKLLTTLRKK FLEGAEIDKS ASVKDEHSTQ 420
421 KHNLRYDLEV IIRSILERYA PIFISLTSNM IEEDLLLQEV TLQRALLDLE NMAKLVSCTP 480
481 MCIWRRSGEI CFVSNEFYSL TGFNKNLLLD RTSFIFEYLD HKSVSNYFQI FNELLAFGYN 540
541 DINKRKKLLM LNACSSTSSK ITEGFSFTTD GKAIFTKCNL LLSNGLYLKC ACCWTVKRDS 600
601 FNIPILVMGQ FLPIFEMD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
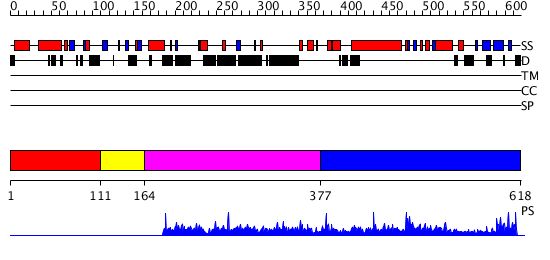
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..110] | 8.05 | No description for 1pyiA was found. | |
| 2 | View Details | [111..163] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [164..376] | 1.003948 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [377..618] | 1.003925 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)