













| General Information: |
|
| Name(s) found: |
MNI1 /
YIL110W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 377 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
response to drug
[IMP |
| Molecular Function: |
S-adenosylmethionine-dependent methyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFSFGFTSN DFDDDELVAQ PETFVESSKE NENTTAYINP LDSDFLSQAG VVQPNVEDLG 60
61 TILESLKDVR LTFEEFQSPI YRKPLIKREL FDVKHQLMLE TDAQSNNNST ELDILLGDTS 120
121 EDLRKNIYEG GLKSWECSYD LVDLLSENVD RISNDIDAVV EIGCGTALPS EFLFRSALLR 180
181 NDRSKGLKFV LTDYNASVLR LVTIPNLVIT WAKTVLTKEQ WYALQKDECE DIPINNEELL 240
241 LTSKLLAAFY DDVQSRNISV TLISGSWGRK FSNLIHEVLS GSQKVLSLSS ETIYQPDNLP 300
301 VIAETILDIH NLPQTDVKTY VAAKDIYFGV GGSITEFEAY LDDKINSEHL PIHSERFKVN 360
361 SGLKRSIICI ETNKAIR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
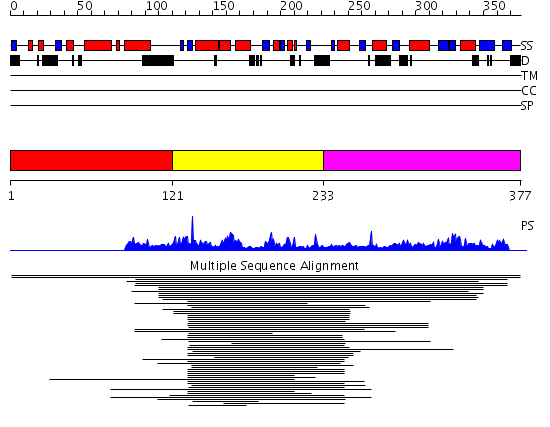
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [121..232] | 2.221849 | Arginine methyltransferase, HMT1 | |
| 3 | View Details | [233..377] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)