













| General Information: |
|
| Name(s) found: |
SHQ1 /
YIL104C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 507 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleoplasm
[IDA |
| Biological Process: |
snoRNA metabolic process
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MITPRFSITQ DEEFIFLKIF ISNIRFSAVG LEIIIQENMI IFHLSPYYLR LRFPHELIDD 60
61 ERSTAQYDSK DECINVKVAK LNKNEYFEDL DLPTKLLARQ GDLAGADALT ENTDAKKTQK 120
121 PLIQEVETDG VSNNIKDDVK TIGQMGEGFN WEIEQKMDSS TNNGILKTKY GFDNLYDTVI 180
181 SVSTSNGNDI NELDDPEHTD ANDRVIERLR KENLKFDPEY YVSEYMTHKY GNEEDLEING 240
241 IKELLKFTPS IVKQYLQWYK DSTNPNLVMP IEFTDEEQKQ MQDNLPKKSY LVEDIKPLYV 300
301 TILSVLFSYV FEQIENEGTH TTESAWTMGK LCPQISFLDQ QLKQVNELQD GMKEISKVNK 360
361 DSSLIKIAII TGIRRALSYP LHRNYDLAMK AWTFVYYILR GGKRLVIRAL LDIHETFRFH 420
421 DVYYVYDKVL LDDLTAWFIS QGSENVIRSL ALEMRKEQES LSKQDIEFEC IASFNEQTGE 480
481 PEWETLNIRE MEILAESEYR EQQQNPQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
ALD6 PTC2 SHQ1 |
| View Details | Ho Y, et al. (2002) |
|
CBF5 PFK2 SHQ1 SRP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Unpublished Data |
| View Data |
|
Huh WK, et al. (2003) |
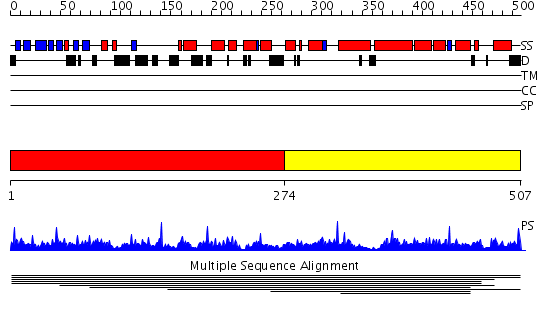
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..273] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [274..507] | 1.007969 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [360..507] | 1.031995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)