













| General Information: |
|
| Name(s) found: |
ETP1 /
YHL010C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 585 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
nuclear localization sequence binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQFEYIITL EFGNQNQVES AYQIFKSIPK KLKAKSIGEE SIKSNNQDWQ DWRVCDLEID 60
61 MITDFKNQTS KEEESDLITS QYLGHGIIRL FKLSNANNTL NEKEILTIPG DDTMICILFV 120
121 PTYFTVHDLL HFYIGDDIVN KQVSNFRILR NQQKGMGFNF TVLIKFRNAL DAKNFKEEFN 180
181 GKSFSRMDPE TCHVISVKEI VFQKKLFQRP AANEDFPYLL TDPFTVKKKK ELVKVELPTC 240
241 PVCLERMDSE TTGLVTIPCQ HTFHCQCLNK WKNSRCPVCR HSSLRLSRES LLKQAGDSAH 300
301 CATCGSTDNL WICLICGNVG CGRYNSKHAI KHYEETLHCF AMDIRTQRVW DYAGDNYVHR 360
361 LVQNEVDGKL VEVGGSGDDD NNDIGNSDEL QNVVYGNRSK NGEKSNSNKK DGELAANFLR 420
421 HREYHLEYVQ VLISQLESQR EYYELKLQEK DQTASDSSNV ESLKKSMEDL KLQFQVTQKE 480
481 WQKREMAQKS KLEEDMLVIE GLQANLDHLS KKQEQLEREN KALEESKQDL EEQVKDLMFY 540
541 LDSQEKFKDA DESVKEGTIL IQQPHGAAQA SKSKKKRNKN KKAGK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
ETP1 MOH1 NHA1 YKR017C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
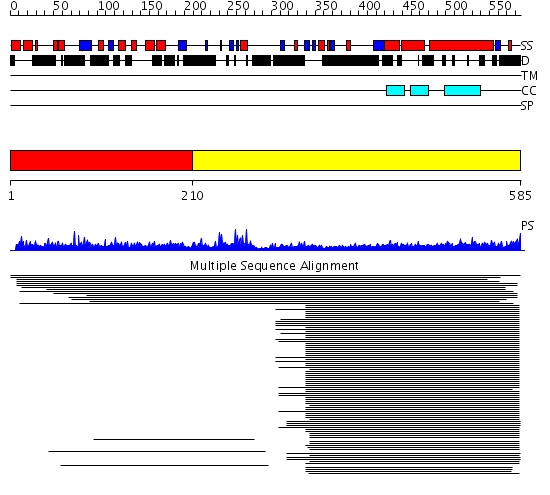
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..209] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [210..585] | 21.09691 | Heavy meromyosin subfragment | |
| 3 | View Details | [189..585] | 24.69897 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)