













| General Information: |
|
| Name(s) found: |
SMI1 /
YGR229C
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 505 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cellular bud tip [TAS |
| Biological Process: |
1,3-beta-glucan biosynthetic process
[TAS cellular cell wall organization [TAS telomere maintenance [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLFKRKVKE WVYSLSTDDH YAEYNPDETP TFNMGKRLNS NNGQVNPSQM HLNSVDEEMS 60
61 MGFQNGVPSN EDINIDEFTS TESNDGVSET LLAWRHIDFW TSEHNPDLNA TLSDPCTQND 120
121 ITHAEEDLEV SFPNPVKASF KIHDGQEDLE SMTGTSGLFY GFQLMTLDQV VAMTQAWRNV 180
181 AKNLNKRSQQ GLSHVTSTGS SSSMERLNGN KFKLPNIPDQ KSIPPNAVQP VYAHPAWIPL 240
241 ITDNAGNHIG VDLAPGPNGK YAQIITFGRD FDTKFVIAEN WGEFLLSFAN DLEAGNWYLV 300
301 DDNDDYFSGD GELVFRDKKS NGPIQDYFEV LKRRTWIKYQ ENLRSQQQKS QPDTSLQEQK 360
361 YVPASQKKVA AEQPSTLNAE SIKGEDSGSA DVQSVQDHES VKIVKTEPSE AETTTVNTES 420
421 LGQAEHEIKA DNVDIKQESE RKEDEKQPKV EEKEHVENEH VTESAKKDDD VNKQTEEMNK 480
481 KEENEIRSDD AKVEEAREEF ENIAL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ANB1 ANR2 GPD2 HYP2 MEF1 MNP1 SMI1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
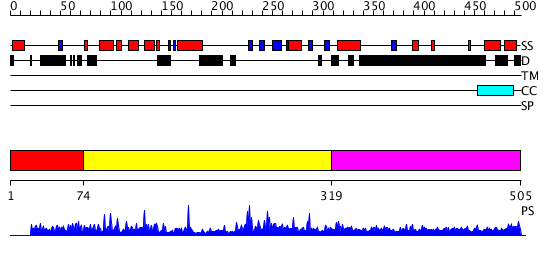
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [74..318] | 1.95 | No description for 2prvA was found. | |
| 3 | View Details | [319..505] | 121.157997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)