













| General Information: |
|
| Name(s) found: |
NMA2 /
YGR010W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 395 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
NAD metabolic process
[TAS |
| Molecular Function: |
nicotinate-nucleotide adenylyltransferase activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPTKAPDFK PPQPNEELQP PPDPTHTIPK SGPIVPYVLA DYNSSIDAPF NLDIYKTLSS 60
61 RKKNANSSNR MDHIPLNTSD FQPLSRDVSS EEESEGQSNG IDATLQDVTM TGNLGVLKSQ 120
121 IADLEEVPHT IVRQARTIED YEFPVHRLTK KLQDPEKLPL IIVACGSFSP ITYLHLRMFE 180
181 MALDDINEQT RFEVVGGYFS PVSDNYQKRG LAPAYHRVRM CELACERTSS WLMVDAWESL 240
241 QSSYTRTAKV LDHFNHEINI KRGGIMTVDG EKMGVKIMLL AGGDLIESMG EPHVWADSDL 300
301 HHILGNYGCL IVERTGSDVR SFLLSHDIMY EHRRNILIIK QLIYNDISST KVRLFIRRGM 360
361 SVQYLLPNSV IRYIQEYNLY INQSEPVKQV LDSKE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
NMA1 NMA2 |
| View Details | Krogan NJ, et al. (2006) |
|
NMA1 NMA2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
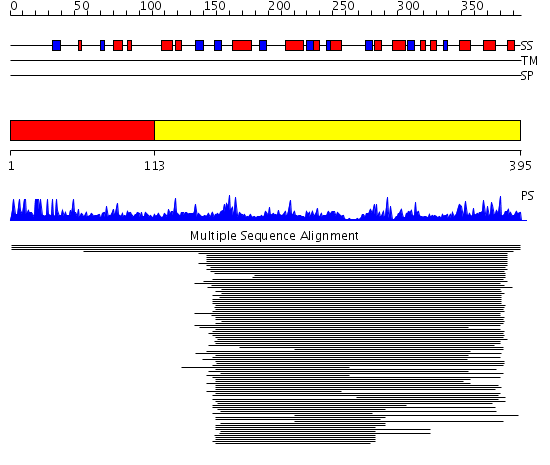
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [113..395] | 604.228787 | Nicotinamide mononucleotide (NMN) adenylyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)