













| General Information: |
|
| Name(s) found: |
YGL082W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 381 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVTFLTKNV QINGTQFKIL LQNGQGECAL IALANVLLIS PAHARYAQEI SRLVRGKETV 60
61 TLNELVQTLA DMGVQNPNGT DVDKQQLLQI LPQLYSGLNI NPEFNGSFED GVEMSIFRLY 120
121 NVGIVHGWII DGDNDPNSYE HVSKYSYMGA QKVLVQSYEI QKNNAQFENS EQIQSDAPYL 180
181 KSFLARSATQ LTEYGLTHLR EILVERSYAV LFRNDHFCTL YKNNGELFTL VTDPTYRNRK 240
241 DINWQSLKSV NGSQDSYYTG NFIPTSLERT ETTATGQNES YISNPFSDQN TGHVTSNQVN 300
301 SGASGVQQIE DDEELARRLQ EQEDMRAANN MQNGYANNGR NHQRERFERP EKNSKKNKFL 360
361 PFNGSNKEKK RDKLKKNCVI M |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
RVS167 YGL082W |
| View Details | Krogan NJ, et al. (2006) |

|
TDP1 YGL082W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | De Craene, J., et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
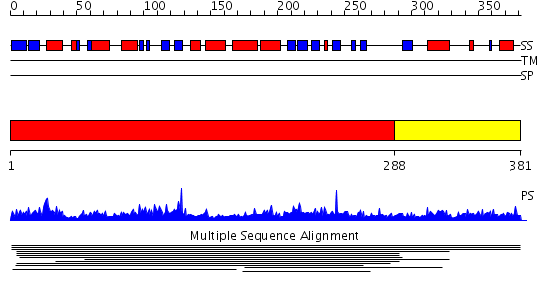
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..287] | 5.011925 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [288..381] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [277..381] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)