













| General Information: |
|
| Name(s) found: |
NPY1 /
YGL067W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 384 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA cytoplasm [IDA |
| Biological Process: |
NADH metabolic process
[IDA |
| Molecular Function: |
NAD+ diphosphatase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTAVTFFGQ HVLNRVSFLR CSKEFIKKSL NHDSTVFIPF IEGEALISPE NGDLVQLSNS 60
61 VKSYKNILSA IVPLYTTLLN TTRSRSDESG INVTFLGLLE GTDSAFNFEW SNISYKGTPY 120
121 FGLDIRVTES TLFKKVDFEP IFSYPKVTRD HIFKQTNEDA SLYSQGKMYL DWLAKYKFCP 180
181 GCGSPLFPVE AGTKLQCSNE NRNVYCNVRD ARINNVCFPR TDPTVIIALT NSDYSKCCLA 240
241 RSKKRYGDFV LYSTIAGFME PSETIEEACI REIWEETGIS CKNIDIVRSQ PWPYPCSLMI 300
301 GCLGIVQFNS KNEVINLNHD DELLDAQWFD TTEIIQALDK YAGGYRVPFK NDINLPGSTT 360
361 IAFQLINHVC ENYKNLRKTS SSHL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
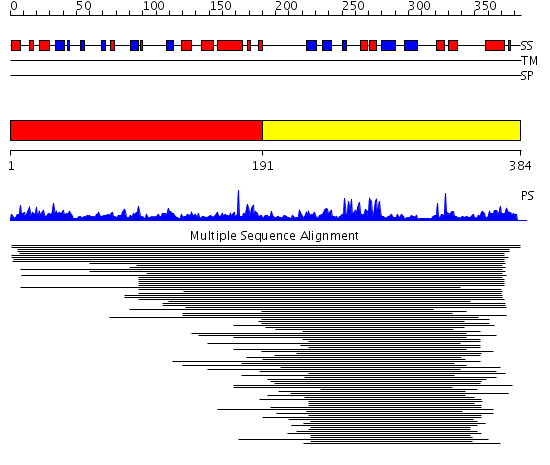
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 1.038996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [191..384] | 16.69897 | ADP-ribose pyrophosphatase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)