













| General Information: |
|
| Name(s) found: |
PUS2 /
YGL063W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 370 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
tRNA modification
[ISS |
| Molecular Function: |
RNA binding
[IEA]
pseudouridine synthase activity [IEA][IGI][IMP] isomerase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLGYCGSGY YGMQYNPPHK TIEGEILTKL FDVGAISEEN SLAPKKNSFM AAARTDKGVH 60
61 AMLNLLSLKI TLREDTVAKL NAALPPEIRV WGIQPVNKKF NARSACDSRW YQYLIPEFIL 120
121 IGPPRSSLLH RNVGGSYRED GSQEVWDTFL EQTRGRFSGD ELCRLQDTAQ KLSESDPLVQ 180
181 DYVGLLSGTL SGYCLSPSKL DAFEAAMQEY VGTHNFHNFT TGKLWGDPSA QRHIKKVVVS 240
241 QASPGWICVR IHGQSFMLHQ IRRMVALAVL AARCQLPPNI VRNYFNAGPR KYIPRAPAQG 300
301 LLLEGPVFDG YNTKLRNLLY CEIRPDDITL ERMCRFRERQ ICTAIAHEET QRHVFCHFVR 360
361 QMNRLATPLI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
COF1 PUS2 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
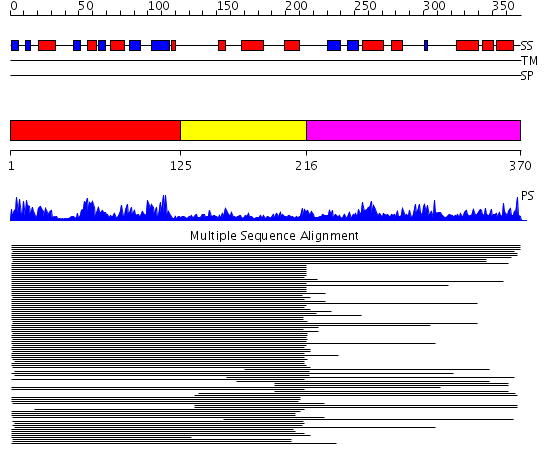
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..124] | 109.980454 | Pseudouridine synthase I | |
| 2 | View Details | [125..215] | 109.980454 | Pseudouridine synthase I | |
| 3 | View Details | [216..370] | 7.920819 | tRNA pseudouridine synthase No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)