













| General Information: |
|
| Name(s) found: |
GEP7 /
YGL057C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 287 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLSNVKIFR LKSHRAFRIG PMIKAVAGNL LVKRFYQPKL ERIPPASLLL KQKIRLAQNG 60
61 STTSTENPIS FSQTMSEIFS VLQPSAPDLD EDETSGLKRD HLLTERLNNG ELGVIMNKFF 120
121 NPSSTHNNQL IDTNILLQNF PKLSGNDLDL LDFAINEKMR GNWNDLKQDF IQLWYYKSFG 180
181 FLGPRTQFVL TNSSPSVRSQ FLKLPFIEYN WFLLQNNKNA NILPADVQNV VKVFHLDDKR 240
241 FTWKSIDPFS KAIISFVVFV SIYVWLDESA KQKTKELPAQ KSTVISE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
GEP7 GIP1 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
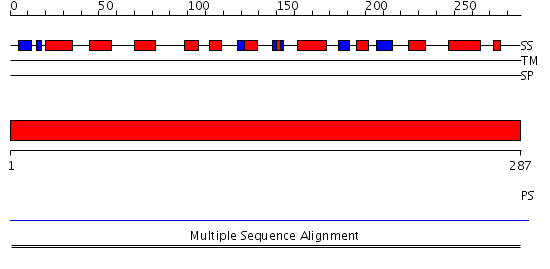
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..287] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [145..287] | 1.35 | Mitochondrial cytochrome c oxidase subunit IV |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)