













| General Information: |
|
| Name(s) found: |
MIG3 /
YER028C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 394 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IGI transcription initiation [TAS response to DNA damage stimulus [IGI |
| Molecular Function: |
DNA binding
[IDA sequence-specific DNA binding [IDA specific transcriptional repressor activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNYLRDRFPP DNDQRPFRCE ICSRGFHRLE HKKRHGRTHT GEKPHKCTVQ GCPKSFSRSD 60
61 ELKRHLRTHT KGVQRRRIKS KGSRKTVVNT ATAAPTTFNE NTGVSLTGIG QSKVPPILIS 120
121 VAQNCDDVNI RNTGNNNGIV ETQAPAILVP VINIPNDPHP IPSSLSTTSI TSIASVYPST 180
181 SPFQYLKSGF PEDPASTPYV HSSGSSLALG ELSSNSSIFS KSRRNLAAMS GPDSLSSSKN 240
241 QSSASLLSQT SHPSKSFSRP PTDLSPLRRI MPSVNTGDME ISRTVSVSSS SSSLTSVTYD 300
301 DTAAKDMGMG IFFDRPPVTQ KACRSNHKYK VNAVSRGRQH ERAQFHISGD DEDSNVHRQE 360
361 SRASNTSPNV SLPPIKSILR QIDNFNSAPS YFSK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
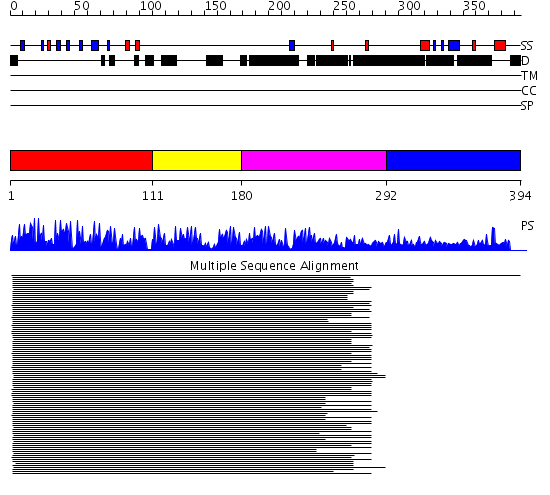
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..110] | 130.522879 | No description for 1lu6A_ was found. | |
| 2 | View Details | [111..179] | 5.0 | Enhancer binding protein | |
| 3 | View Details | [180..291] | 3.104994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [292..394] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)