













| General Information: |
|
| Name(s) found: |
DLD3 /
YEL071W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 496 amino acids |
Gene Ontology: |
|
| Cellular Component: |
soluble fraction
[IDA cytoplasm [IDA |
| Biological Process: |
lactate metabolic process
[ISS |
| Molecular Function: |
D-lactate dehydrogenase (cytochrome) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAAHPVAQL TAEAYPKVKR NPNFKVLDSE DLAYFRSILS NDEILNSQAP EELASFNQDW 60
61 MKKYRGQSNL ILLPNSTDKV SKIMKYCNDK KLAVVPQGGN TDLVGASVPV FDEIVLSLRN 120
121 MNKVRDFDPV SGTFKCDAGV VMRDAHQFLH DHDHIFPLDL PSRNNCQVGG VVSTNAGGLN 180
181 FLRYGSLHGN VLGLEVVLPN GEIISNINAL RKDNTGYDLK QLFIGAEGTI GVVTGVSIVA 240
241 AAKPKALNAV FFGIENFDTV QKLFVKAKSE LSEILSAFEF MDRGSIECTI EYLKDLPFPL 300
301 ENQHNFYVLI ETSGSNKRHD DEKLTAFLKD TTDSKLISEG MMAKDKADFD RLWTWRKSVP 360
361 TACNSYGGMY KYDMSLQLKD LYSVSAAVTE RLNAAGLIGD APKPVVKSCG YGHVGDGNIH 420
421 LNIAVREFTK QIEDLLEPFV YEYIASKKGS ISAEHGIGFH KKGKLHYTRS DIEIRFMKDI 480
481 KNHYDPNGIL NPYKYI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
ACS2 ATG19 CAR2 DLD3 DUG1 ERG10 FUM1 ILV3 LAP4 RHO1 WTM1 |
| View Details | Qiu et al. (2008) |

|
DLD3 LEU1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
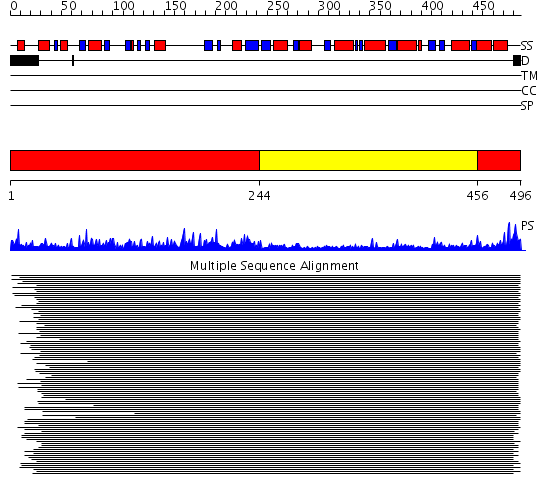
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..243] [456..496] |
115.38764 | Flavoprotein subunit of p-cresol methylhydroxylase | |
| 2 | View Details | [244..455] | 115.38764 | Flavoprotein subunit of p-cresol methylhydroxylase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)