













| General Information: |
|
| Name(s) found: |
MMS21 /
YEL019C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 267 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA nucleus [IDA cytoplasm [IDA Smc5-Smc6 complex [IPI |
| Biological Process: |
DNA repair
[IMP |
| Molecular Function: |
SUMO ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALNDNPIPK SVPLHPKSGK YFHNLHARDL SNIYQQCYKQ IDETINQLVD STSPSTIGIE 60
61 EQVADITSTY KLLSTYESES NSFDEHIKDL KKNFKQSSDA CPQIDLSTWD KYRTGELTAP 120
121 KLSELYLNMP TPEPATMVNN TDTLKILKVL PYIWNDPTCV IPDLQNPADE DDLQIEGGKI 180
181 ELTCPITCKP YEAPLISRKC NHVFDRDGIQ NYLQGYTTRD CPQAACSQVV SMRDFVRDPI 240
241 MELRCKIAKM KESQEQDKRS SQAIDVL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
MMS21 NSE4 |
| View Details | Hazbun TR, et al. (2003) |
|
KRE29 MMS21 NSE1 NSE3 NSE4 NSE5 SMC5 SMC6 |
| View Details | Krogan NJ, et al. (2006) |
|
GDH2 MMS21 NSE3 NSE4 SMC5 YOR352W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
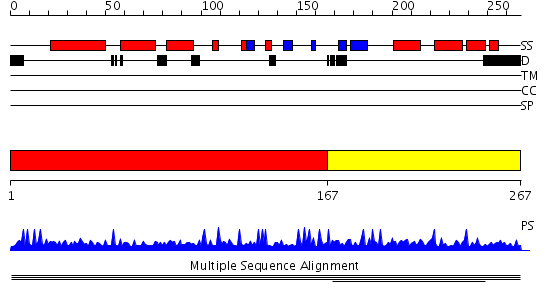
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [167..267] | 3.065502 | MIZ zinc finger No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)