













| General Information: |
|
| Name(s) found: |
UBP1 /
YDL122W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IC endoplasmic reticulum [IDA |
| Biological Process: |
protein deubiquitination
[TAS |
| Molecular Function: |
ubiquitin-specific protease activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLFIESKIN SLLQFLFGSR QDFLRNFKTW SNNNNNLSIY LLIFGIVVFF YKKPDHLNYI 60
61 VESVSEMTTN FRNNNSLSRW LPRSKFTHLD EEILKRGGFI AGLVNDGNTC FMNSVLQSLA 120
121 SSRELMEFLD NNVIRTYEEI EQNEHNEEGN GQESAQDEAT HKKNTRKGGK VYGKHKKKLN 180
181 RKSSSKEDEE KSQEPDITFS VALRDLLSAL NAKYYRDKPY FKTNSLLKAM SKSPRKNILL 240
241 GYDQEDAQEF FQNILAELES NVKSLNTEKL DTTPVAKSEL PDDALVGQLN LGEVGTVYIP 300
301 TEQIDPNSIL HDKSIQNFTP FKLMTPLDGI TAERIGCLQC GENGGIRYSV FSGLSLNLPN 360
361 ENIGSTLKLS QLLSDWSKPE IIEGVECNRC ALTAAHSHLF GQLKEFEKKP EGSIPEKLIN 420
421 AVKDRVHQIE EVLAKPVIDD EDYKKLHTAN MVRKCSKSKQ ILISRPPPLL SIHINRSVFD 480
481 PRTYMIRKNN SKVLFKSRLN LAPWCCDINE INLDARLPMS KKEKAAQQDS SEDENIGGEY 540
541 YTKLHERFEQ EFEDSEEEKE YDDAEGNYAS HYNHTKDISN YDPLNGEVDG VTSDDEDEYI 600
601 EETDALGNTI KKRIIEHSDV ENENVKDNEE LQEIDNVSLD EPKINVEDQL ETSSDEEDVI 660
661 PAPPINYARS FSTVPATPLT YSLRSVIVHY GTHNYGHYIA FRKYRGCWWR ISDETVYVVD 720
721 EAEVLSTPGV FMLFYEYDFD EETGKMKDDL EAIQSNNEED DEKEQEQKGV QEPKESQEQG 780
781 EGEEQEEGQE QMKFERTEDH RDISGKDVN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
DRS1 ERB1 HAA1 HRQ1 MKT1 STE20 UBP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
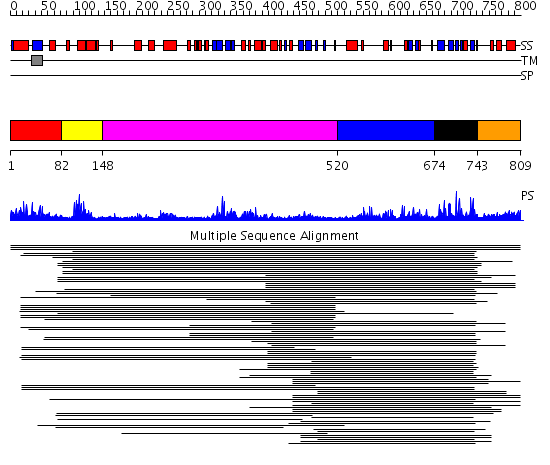
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [82..147] | 12.619789 | No description for PF00442 was found. No confident structure predictions are available. | |
| 3 | View Details | [148..519] | 2.134989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [520..673] | 2.171993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [674..742] | 22.119186 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. | |
| 6 | View Details | [743..809] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)