













| General Information: |
|
| Name(s) found: |
RDS1 /
YCR106W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 832 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
response to xenobiotic stimulus
[IMP |
| Molecular Function: |
DNA binding
[IPI transcription factor activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSITVKKPR LRLVCLQCKK IKRKCDKLRP ACSRCQQNSL QCEYEERTDL SANVAANDSD 60
61 GFNSSHKLNF EQQPVLERTG LRYSLQVPEG VVNATLSIWN AEDMLVIVGL VTFLDYPFAA 120
121 HSLAQHDQYI RALCASLYGM ALVDFSNYAN GIPCEDTSRS ILGPLSFIEK AIFRRIEHSK 180
181 QFRVQSAALG LLYNAFSMEE ENFSTLLPSL IAEVEDVLMQ KKDCEILLRC FYQNIYPFYP 240
241 FMDISLFESD LTSLLLQDDN NRWKISTEVK NVRKKIETLS LLTIVMAMAL MHSKLDANLL 300
301 SMVKENASES ARKLSLLCHK LLCLLDVFRY PNENTFTCLL YFYVSEHLDP ESPDCVLSPT 360
361 NLLTLHHLLN LSMTLGLQYE PSKYKRFKDP EVIRQRRILW LGVQSLLFQI SLAEGDAGKS 420
421 NSEYMEAYLT DFEEYIEASS EYEKSSASES NVQMNDIVWN KYKFHVILSK LMSDCTSVIQ 480
481 HPQLFHILGN IKRSEDFMAE NFPTSSIYQP LHEKEPNAIK VGKSTVLDVM DIQKTEIFLT 540
541 NIVGSMCFLN IFDVLSLHFE KKCVMHWEEY EKNYHFLTLK SFNAYLKLAG LISDYLENKF 600
601 QGNILESRGY IIDKQICFML VRIWMFQCRI LLRFSYKQES QKKLASSSIS TNDNEKEDEM 660
661 IVILERLIKH IRNQMAHLVD LAKGKLQDSY FGAYQTVPMF RYVVYLIDVG GLVSVTNGFW 720
721 DKISSDGEIP PKVQQAVRLK WGLDCNNSRR IKQKLISSQS LQSFNQVLLC QMEDAVLSSS 780
781 FAIKANTAMS QNTAEEFFNI SEEEALNQLL ENNNFDAFWD LLGENLSDMP SL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
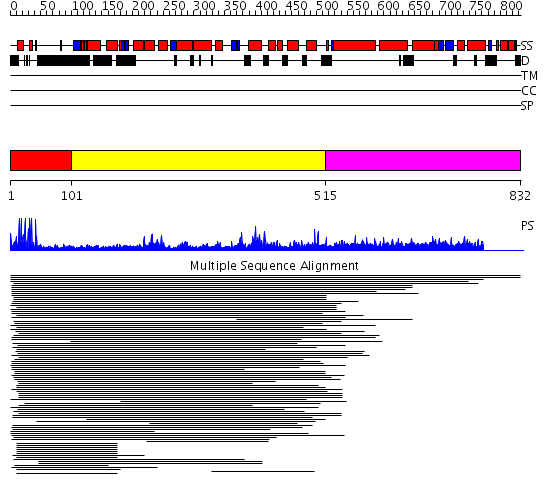
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | 49.38764 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [101..514] | 1.131971 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [515..832] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)