













| General Information: |
|
| Name(s) found: |
SMY2 /
YBR172C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 790 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
cytoskeleton organization
[TAS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLLELFTIL QIPTHAVQWS LRVHHFHSYS LSFYLPSSSP IYSTSQPTSI MIAPDSQRLF 60
61 GSFDEQFKDL KLDSVDTENN NTHGVSTILD SSPASVNNNT NGAVAASVNT VPGSTFRSNT 120
121 PLLGGRHPLS RTSSLIDSIG IQRAASPFSS MKEPFIPQSS GVMSSSFWHG DHPESRVSTP 180
181 VQQHPLLQRN ESSSSFSYAA NLGVNLSTHS LAVDITPLST PTAAQSHVNL FPSSDIPPNM 240
241 SMNGMSQLPA PVSVESSWRY IDTQGQIHGP FTTQMMSQWY IGGYFASTLQ ISRLGSTPET 300
301 LGINDIFITL GELMTKLEKY DTDPFTTFDK LHVQTTSSDS INLNLAPYAS GVAATGTIKA 360
361 TENDIFKPLT HDNIWDMDGG TTSKGVDIKL ASATTISQTD ESHKQEYKST TMLEKGKKEK 420
421 SESVAKALLD EQEKRNRELK RKEEARLSKK QKQKEDDLLK KQKEQKEQKE KEALEAEKQK 480
481 KSEKTKKDTQ TQTEGFKTSK DLPSLNSSSA NPAPWASKVK VNNAIETSIK NGVSSTGKKK 540
541 GEPLGLQQRN SKEEKQKEEL KSVLNWANKS SLPSNQTIDI KSQFQKSPKG MKESSPLKEL 600
601 EDPNFIEEQK KLWEKVQSSS KQVKSTSSAS TTTSSWTTVT SKGKAPIGTV VSPYSKTNTS 660
661 LNSSLTAKTS TTSTTTTFAS MNNVSPRQEF IKWCKSQMKL NSGITNNNVL ELLLSLPTGP 720
721 ESKELIQETI YANSDVMDGR RFATEFIKRR VACEKQGDDP LSWNEALALS GNDDDGWEFQ 780
781 VVSKKKGRKH |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
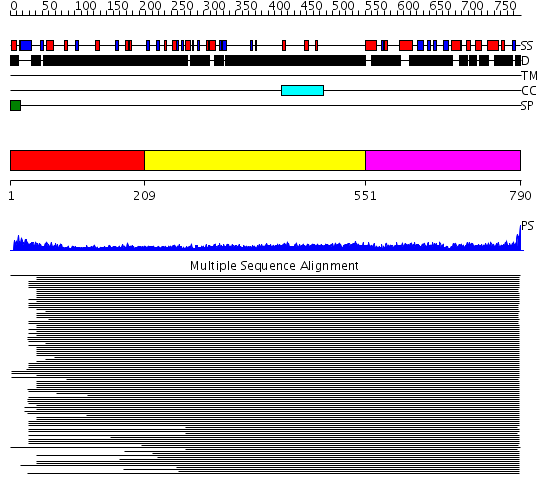
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..208] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [209..550] | 20.0 | Heavy meromyosin subfragment | |
| 3 | View Details | [551..790] | 10.519997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)