













| General Information: |
|
| Name(s) found: |
IML3 /
YBR107C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 245 amino acids |
Gene Ontology: |
|
| Cellular Component: |
outer kinetochore of condensed nuclear chromosome
[IDA |
| Biological Process: |
ascospore formation
[IMP meiotic sister chromatid segregation [IMP chromosome segregation [TAS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPYTWKFLGI SKQLSLENGI AKLNQLLNLE VDLDIQTIRV PSDPDGGTAA DEYIRYEMRL 60
61 DISNLDEGTY SKFIFLGNSK MEVPMFLCYC GTDNRNEVVL QWLKAEYGVI MWPIKFEQKT 120
121 MIKLADASIV HVTKENIEQI TWFSSKLYFE PETQDKNLRQ FSIEIPRESC EGLALGYGNT 180
181 MHPYNDAIVP YIYNETGMAV ERLPLTSVIL AGHTKIMRES IVTSTRSLRN RVLAVVLQSI 240
241 QFTSE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
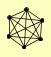
|
AME1 CHL4 CTF19 IML3 MCM22 NKP2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
CHL4 IML3 |
| View Details | Krogan NJ, et al. (2006) |
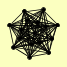
|
AME1 ARG5,6 CHL4 CTF19 CTF3 IML3 MCM16 MCM21 MCM22 NKP1 NKP2 OKP1 PRP18 SET4 YNR062C |
| View Details | Gavin AC, et al. (2002) |
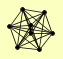
|
AME1 CHL4 CTF19 CTF3 IML3 MCM21 MCM22 NKP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
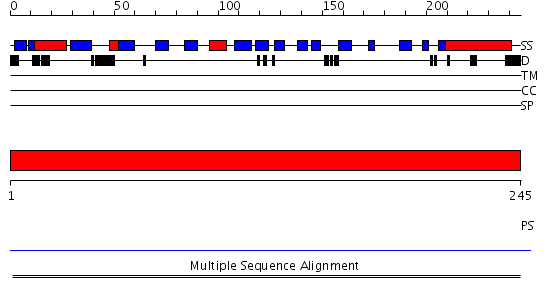
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..245] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)