













| General Information: |
|
| Name(s) found: |
rdx-PE /
FBpp0099767
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 829 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
eye development
[IMP]
eye morphogenesis [IMP] positive regulation of JNK cascade [IMP] positive regulation of apoptosis [IGI] regulation of proteolysis [IMP] establishment of ommatidial planar polarity [IMP] segment polarity determination [IMP] cellularization [IMP] protein ubiquitination [IMP] |
| Molecular Function: |
protein binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFEPYVKIKK KRSVNEIYEN ENLEQQQHHL QQQQQQPATS DNCCCENGEP QNAPEVATAT 60
61 VAATSVAATS AAATVATGAA ASSSSSGNCS RLQQLISTPP VLLRRSSLQQ QQHQQQQHHP 120
121 HTPAATATPP QQQQQQQAAP SVLQQHLGHL NYESGATAAA AATAAAAAAT VATSRSGSAT 180
181 LAQHLATPSN ILQAAFGSSN LQHILTRSAP SPSSSAISSN NCSSACAGNT HYNGGNSNSG 240
241 SSSSNSNHHS NSIIASRLFG AASSSSSSSS SSASASSSSV AASSSSSSHH LHSHHSALTN 300
301 SITNRINQSI RRHLNQQQHH HPLSASSSSA SASPSASTSS SSSYQQSSVQ QQHYNCAHPA 360
361 QQQQHHHHHH SSSSSSSSSS SSSHHHHNSS SSSNSNNQQQ PQQSPLCLVL LVKCPNSKEF 420
421 CNAAANFCDK RLPVNECQAS QTARVTSNLH ASSSTMAVSR VPSPPLPEVN TPVAENWCYT 480
481 QVKVVKFSYM WTINNFSFCR EEMGEVLKSS TFSAGANDKL KWCLRVNPKG LDEESKDYLS 540
541 LYLLLVSCNK SEVRAKFKFS ILNAKREETK AMESQRAYRF VQGKDWGFKK FIRRDFLLDE 600
601 ANGLLPEDKL TIFCEVSVVA DSVNISGQSN IVQFKVPECK LSEDLGNLFD NEKFSDVTLS 660
661 VGGREFQAHK AILAARSDVF AAMFEHEMEE RKLNRVAITD VDHEVLKEML RFIYTGKAPN 720
721 LEKMADDLLA AADKYALEKL KVMCEEALCV NLSVETAAET LILADLHSAD QLKAQTIDFI 780
781 NTHATDVMET SGWQNMITTH SHLIAEAFRA LATQQIPPIG PPRKRVKMS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
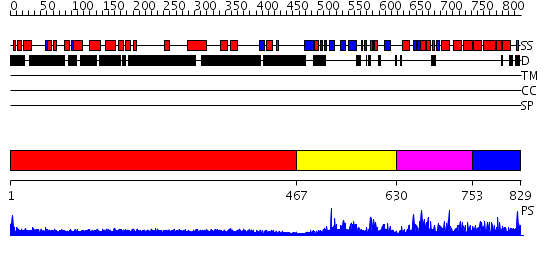
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..466] | 4.045757 | No description for 1y0fB was found. | |
| 2 | View Details | [467..629] | 45.154902 | Solution structure of N-terminal domain of speckle-type POZ protein | |
| 3 | View Details | [630..752] | 30.0 | No description for 2nn2A was found. | |
| 4 | View Details | [753..829] | 8.0 | No description for 2eqxA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)