













| General Information: |
|
| Name(s) found: |
Y3797_VIBC3
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Vibrio cholerae O395 |
| Length: | 224 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLKQLPTES MPREKLLQRG PQSLSDAELL AIFLRTGTQG MNVLALADLL LRDFGSLRAL 60
61 FCASKEQFCR HKGLGEAKFV QLQAVLEMTQ RYLAETLKRG DALTSPQQTK LYLSSVLRDR 120
121 QREAFYILFL DNQHRVIRDE ILFEGTIDAA SVYPREVVKR ALHHNAAAVI LAHNHPSGVA 180
181 EPSQADRRIT DRLRDALGLV EIRVLDHFVV GDGEVVSFAE RGWI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
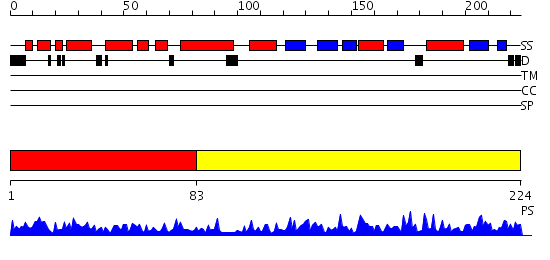
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 9.0 | Crystal Structure Of Archaeal Xpf/Mus81 Homolog, Hef From Pyrococcus Furiosus, Helix-hairpin-helix Domain | |
| 2 | View Details | [83..224] | 40.69897 | No description for 2qlcA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)