













| General Information: |
|
| Name(s) found: |
daf-16 /
CE38722
[WormBase]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 487 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
response to heat
[IEP dauer larval development [IGI insulin receptor signaling pathway [IDA determination of adult lifespan [IGI regulation of transcription, DNA-dependent [IEA positive regulation of transcription from RNA polymerase II promoter [IDA |
| Molecular Function: |
transcription factor activity
[IDA protein binding [IPI sequence-specific DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFSTDFDDD FFNLDLHQQE RSASFGGVTQ YSQQFLREKC SFSPYFHTSL ETVDSGRTSL 60
61 YGSNEQCGQL GGASSNGSTA MLHTPDGSNS HQTSFPSDFR MSESPDDTVS GKKTTTRRNA 120
121 WGNMSYAELI TTAIMASPEK RLTLAQVYEW MVQNVPYFRD KGDSNSSAGW KNSIRHNLSL 180
181 HSRFMRIQNE GAGKSSWWVI NPDAKPGRNP RRTRERSNTI ETTTKAQLEK SRRGAKKRIK 240
241 ERALMGSLHS TLNGNSIAGS IQTISHDLYD DDSMQGAFDN VPSSFRPRTQ SNLSIPGSSS 300
301 RVSPAIGSDI YDDLEFPSWV GESVPAIPSD IVDRTDQMRI DATTHIGGVQ IKQESKPIKT 360
361 EPIAPPPSYH ELNSVRGSCA QNPLLRNPIV PSTNFKPMPL PGAYGNYQNG GITPINWLST 420
421 SNSSPLPGIQ SCGIVAAQHT VASSSALPID LENLTLPDQP LMDTMDVDAL IRHELSQAGG 480
481 QHIHFDL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
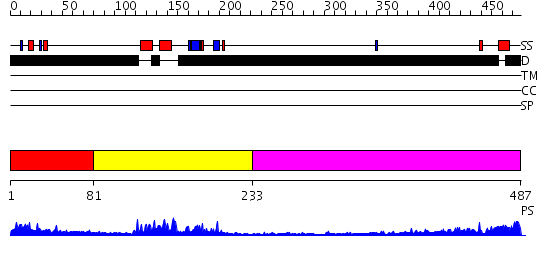
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | 1.358996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [81..232] | 25.045757 | Afx (Foxo4) | |
| 3 | View Details | [233..487] | 5.458996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)