













| General Information: |
|
| Name(s) found: |
gi|34098813
[NCBI NR]
gi|22329895 [NCBI NR] gi|20466614 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 776 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intrinsic to membrane
[IEA]
|
| Biological Process: |
signal transduction
[IEA]
innate immune response [IEA] apoptosis [IEA] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA] transmembrane receptor activity [IEA] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSSSSHNW LYDVFLSFRG EDVRVTFRSH FLKELDRKLI TAFRDNEIER SHSLWPDLEQ 60
61 AIKESRIAVV VFSKNYASSS WCLNELLEIV NCNDKIVIPV FYHVDPSQVR HQIGDFGKIF 120
121 ENTCKRQTDE EVKNQWKKAL TLVANMLGFD SAKWNDEAKM IEEIANDVLG KLLLTTPKDS 180
181 EELVGIEDHI AEMSLLLQLE SKEVRMVGIS GSSGIGKTTI ARALFKRLSR HFQGSTFIDR 240
241 AFVSYSRNIY SGANPDDPNM KLQLQGHFLS EILGKKDIKI DDPAALEERL KHQKVLIIID 300
301 DLDDIMVLDT LVGQTQWFGY GSRIIVVTND KHFLIAHGID HIYEVSFPTD VHACQMLCQS 360
361 AFKQNYAPKG FEDLVVDVVR HAGNFPLGLN LLGKYLRRRD MEYWMDMLPR LENSLRIDGK 420
421 IEKILRISYD GLESEDQEIF RHIACLFNHM EVTTIKSLLA DSDVSFALEN LADKSLIHVR 480
481 QGYVVMHRSL QEMGRKIVRI QSIDKPGERE FLVDPNDIHD ILNACTGTQK VLGISLDIRN 540
541 IRELDVHERA FKGMSNLRFL EIKNFGLKED GLHLPPSFDY LPRTLKLLCW SKFPMRCMPF 600
601 GFRPENLVKL EMQYSKLHKL WEGVAPLTCL KEMDLHGSSN LKVIPDLSEA TNLEILNLKF 660
661 CESLVELPSS IRNLNKLLNL DMLNCKSLKI LPTGFNLKSL DRLNLYHCSK LKTFPKFSTN 720
721 ISVLNLNLTN IEDFPSNLHL ENLVEFRISK EESDEKQWEE EKVSSSTLNI ISKLFP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
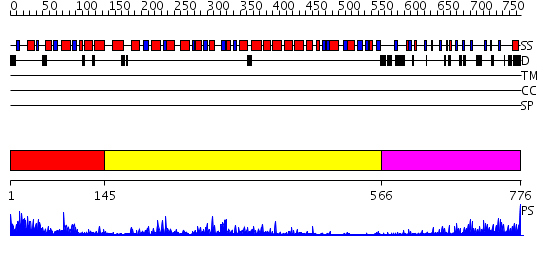
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | 4.31 | Toll-like receptor 2, TLR2 | |
| 2 | View Details | [145..565] | 9.522879 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [566..776] | 6.69897 | No description for 2z7xA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)