













| General Information: |
|
| Name(s) found: |
HGNC:3650|MIM:600393|Ensembl:ENSG00000168496|HPRD:02670,HsxXG003567,G03,HsCD00305507,272624.0,pANT7_cGST,FUSION,NM_004111,2237.0,FEN1,FEN-1|MF1|RAD2,flap
[120926-122011-mus-fusion.fasta]
|
| Description(s) found: |
|
| Organism: | SPECIES UNKNOWN |
| Length: | 380 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIQGLAKLI ADVAPSAIRE NDIKSYFGRK VAIDASMSIY QFLIAVRQGG DVLQNEEGET 60
61 TSHLMGMFYR TIRMMENGIK PVYVFDGKPP QLKSGELAKR SERRAEAEKQ LQQAQAAGAE 120
121 QEVEKFTKRL VKVTKQHNDE CKHLLSLMGI PYLDAPSEAE ASCAALVKAG KVYAAATEDM 180
181 DCLTFGSPVL MRHLTASEAK KLPIQEFHLS RILQELGLNQ EQFVDLCILL GSDYCESIRG 240
241 IGPKRAVDLI QKHKSIEEIV RRLDPNKYPV PENWLHKEAH QLFLEPEVLD PESVELKWSE 300
301 PNEEELIKFM CGEKQFSEER IRSGVKRLSK SRQGSTQGRL DDFFKVTGSL SSAKRKEPEP 360
361 KGSTKKKAKT GAAGKFKRGK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
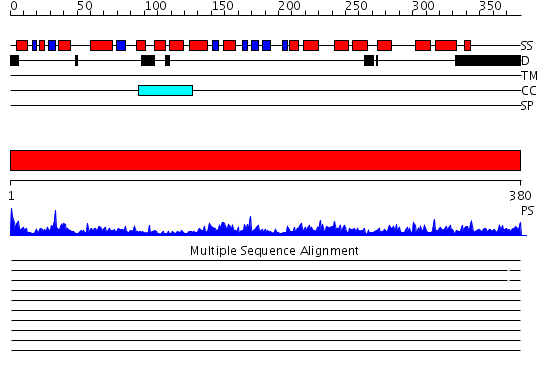
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..380] | 112.0 | Crystal structure of the human FEN1-PCNA complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)