













| General Information: |
|
| Name(s) found: |
Y163_SYNY3
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Synechocystis sp. PCC 6803 substr. Kazusa |
| Length: | 1693 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVMLSCLTQ QQELLQSLAD RVGQPAIQDN LYFCLAGQFT WVEDWYTTAI AGDVLIFDAG 60
61 EKEQLARVLQ QWEQFTGHVI LITGLTEEPD PQTFLTQLAL GKQQRCGERH FHWIVLVDPT 120
121 IEEAFRGLQT FLDGKSHWLD LRLGVGDLEQ CTDELWQCVW LEPSPLAPEK RQEMLVNLPH 180
181 WLAEIVDQKQ NLELEPISLL QAKLSLLQGL ARESRGEMGR KQAQCHYRES LDVWELLGDT 240
241 EAIIWLSLRL GYLFLLQAYG EKSRGHQLWQ QTRNYAQTAI AALENQQWHF THGETLNLLG 300
301 EILRGLEDWE QLRQAAENSL IFFYQLSPFA ATDTTPERNQ EKLWTELELQ GLISLAHSYL 360
361 CEALVEQWKF DEGKEALKRA FETRPKQVEE RDYRPCLANL HYLAGRVQLA NDQIRESLMT 420
421 LRQAQTLVSF EDNPRLYLAI LVELRECYLQ LEDWLAALAI DQEYQAREYR LGQRAFIGPK 480
481 PLPCWPEQRI CRHPLAPEKM LGGGAEARSA VQSKTMVSLT WQDLQKVWEQ KPTPVLVLTG 540
541 EPGVGKTSWL AGEVLQQMSP ERVLLVDFTP HWAEKLWVHL GESFSLPSLG QATTPELVSA 600
601 LQALPILDLL LVLDGENGSL PWQKSPSLRQ QELAQVLWQW LLTTSPSQGV RLLISVPPTA 660
661 IADLYEELKG QLGEEHSLPP LQYQTLSPPT LGQAENWLTQ AMAQCRHPWP PSLQSQFLGD 720
721 LAMEGNGEQQ PWLHPIDLQL LGTVLEQEQV TKSADYQGKK LDEWLTLAVQ TYLAFLPPHL 780
781 LKKALQLLKS LADGQQALCL KTEYQLLAAL YSAPEPAADN PPEFAQEDIQ QLQLLLTLLL 840
841 RGRLIIVISQ ATLPYYRLST THLAHALQGK SAVISTPALI NAGRRGSKSN PNLIAPSPGD 900
901 RQQGNEDLIA ELEAGSSAEE QVVAQKLKAA QLQYQKLVAG INLEKQCQFI LKQFLVYPLE 960
961 ALLAAVKTGQ DLQKLVTPET PLVQYPSLAP WLSLHSILAR INECNRCHHE GPVTVLRISP1020
1021 SMENTPPLVL TATTNGIAYL WSFHGELINV LRGHQEAITA LDWSADGQYF ATASADHTVK1080
1081 LWQRHGEEVA TLRGHEDWVR SVHFSPHHQF LVTSGQDNTA RIWNFAGEQL TLCQGHADWV1140
1141 RNAEFNCHGQ ILLTASRDGT ARLWDLEGRE IGLCQGHTSW VRNAQFSPDG QWIVTCSADG1200
1201 TARLWDLSSQ CFAVLKGHQN WVNNALWSPD GQHIITSSSD GTARVWSRHG KCLGTLRGHD1260
1261 HNIHGARFSL DGQKIVTYST DNTARLWTKE GTLLTILRGH QKEVYDADFS ADGRFVFTVS1320
1321 ADQTARQWDI SQKDTITLTG HSHWVRNAHF NPKGDRLLTV SRDKTARLWT TEGECVAVLA1380
1381 DHQGWVREGQ FSPDGQWIVT GSADKTAQLW NVLGKKLTVL RGHQDAVLNV RFSPDSQYIV1440
1441 TASKDGTARV WNNTGRELAV LRHYEKNIFA AEFSADGQFI VTASDDNTAG IWEIVGREVG1500
1501 ICRGHEGPVY FAQFSADSRY ILTASVDNTA RIWDFLGRPL LTLAGHQSIV YQARFSPEGN1560
1561 LIATVSADHT ARLWDRSGKT VAVLYGHQGL VGTVDWSPDG QMLVTASNDG TARLWDLSGR1620
1621 ELLTLEGHGN WVRSAEFSPD GRWVLTSSAD GTAKLWPVKT LPQLLSQGGQ WLKNYLTHNA1680
1681 LVSPADRPGA KVT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
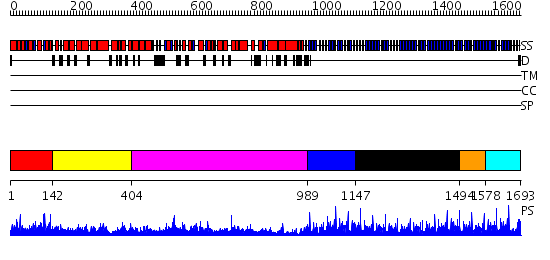
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [142..403] | 25.045757 | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | |
| 3 | View Details | [404..988] | 51.522879 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 4 | View Details | [989..1146] | 72.69897 | No description for 2ovpB was found. | |
| 5 | View Details | [1147..1493] | 3.39794 | No description for 2gbcA was found. | |
| 6 | View Details | [1494..1577] | 3.79 | Crystal Structure Of The C-Terminal WD40 Domain Of Sif2 | |
| 7 | View Details | [1578..1693] | 84.69897 | No description for 2gnqA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)