













| General Information: |
|
| Name(s) found: |
gi|24374196
[NCBI NR]
gi|24348709 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 318 amino acids |
Gene Ontology: |
|
| Cellular Component: |
provirus
[TAS |
| Biological Process: |
transposition, DNA-mediated
[ISS |
| Molecular Function: |
transposase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNVHSLTAK DSKAEIRKTD VVMQINTLIE AGRVVAAHLA QEISVAPSTL SQVLNGQYKA 60
61 NTTQVVKKLD NWLRMREQRE ANPNLDPGFV LTPTANLVMD NLSYAQITQA IVVIIGASGV 120
121 GKSRALEEYQ RSNNNVWKVI ASPSRSSLTE LLYEIAMELG MAQAPRTKGP LARVIRQRLK 180
181 GSEGLIIVDE ADHLDYPTLE ELRILQEETG IGMALVGNNK VYTQLTGGRR NEDYARLFSR 240
241 IAKKLSINKT KQADVRAIAK AWNLQHDAEI SLMIQISEKP GGLRLLSKTL KLAAMFAKGA 300
301 AITEALLRTA FNELETNE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
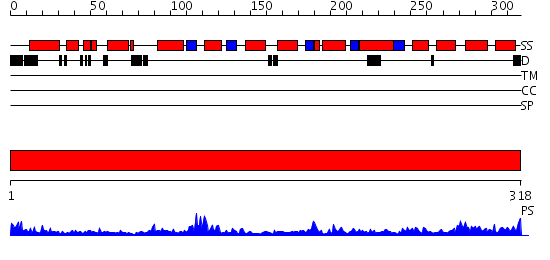
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..318] | 50.522879 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)