













| General Information: |
|
| Name(s) found: |
VMS1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 632 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSQKASKMT GSLKKNDLYI FDLSEQLLNS LKLMSFDSTL REVEVEKTSD NDRNKESGDL 60
61 QIARKKVTSN VMRCSVCQMS FDSRNEQKAH YQTDYHLMNV KRNLRGLDIL SVEEFDALIS 120
121 KEHGIKSEDE NSGGEQTSSD HEESEEASDR DPDLQTNNYM ETIIENDLQK LGFQKDESDA 180
181 ISHINTQSPY IYFKSKYLQK NEVLAIYKSL FNKRSLSNPN EALTFWNSQE NPMAISALFM 240
241 VGGGHFAGAI VSHQRLNVKG NAHKKDETLI EQAVNFLEHK TFHRYTTRRK QGGSQSAMDN 300
301 AKGKANSAGS ALRRYNESAL KTDIQGVLKD WEPYLSKCDN IFIRARNVSD KKIFTDNTVL 360
361 NKGDERIKSF PFTTNRPTVL ELKKAWCELS YLKILPKPEP LAVKETVQKL EVSNKKDEFK 420
421 EKQEPLLEEI QTEEIISLLK KGRAPLLISF LKKNKLDGNF RLKPESKYSL TPTMLHYASQ 480
481 QGMKQMALIL LSNIKCDPTI KNRLGRTAWD LNRNDDVRHA FQIARYNLGE SFTNWDETHI 540
541 GQPLSREQVD EINEKKKAIE NEKAEKLIKL ELEAAKEKQR FAKDAERGPG KKLTNIPSIQ 600
601 QQNLNSLTDE QRRRLMREQR ARAAEERMKK KY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
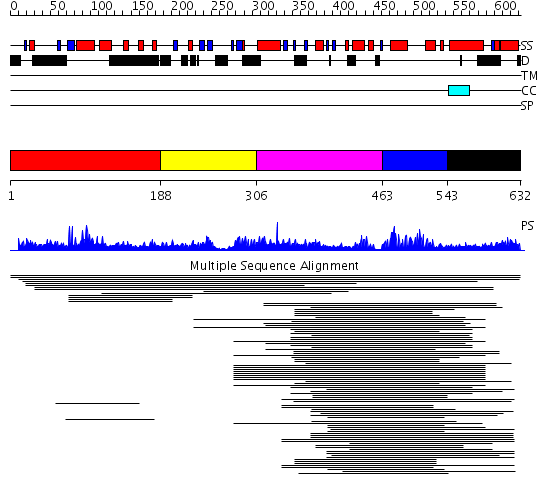
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..187] | 11.021989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [188..305] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [306..462] | 20.522879 | I-kappa-B-alpha | |
| 4 | View Details | [463..542] | 20.522879 | I-kappa-B-alpha | |
| 5 | View Details | [543..632] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)