













| General Information: |
|
| Name(s) found: |
SPT6_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1451 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEETGDSKLV PRDEEEIVND NDETKAPSEE EEGEDVFDSS EEDEDIDEDE DEARKVQEGF 60
61 IVNDDDENED PGTSISKKRR KHKRREREED DRLSEDDLDL LMENAGVERT KASSSSGKFK 120
121 RLKRVGDEGN AAESESDNVA ASRQDSTSKL EDFFSEDEEE EESGLRNGRN NEYGRDEEDH 180
181 ENRNRTADKG GILDELDDFI EDDEFSDEDD ETRQRRIQEK KLLREQSIKQ PTQITGLSSD 240
241 KIDEMYDIFG DGHDYDWALE IENEELENGN DNNEAEEEEI DEETGAIKST KKKISLQDIY 300
301 DLEDLKKNLM TEGDMKIRKT DIPERYQELR AGITDYGNMS SEDQELERNW IAEKISVDKN 360
361 FDANYDLTEF KEAIGNAIKF ITKENLEVPF IYAYRRNYIS SREKDGFLLT EDDLWDIVSL 420
421 DIEFHSLVNK KDYVQRFYAE LHIDDPIVTE YFKNQNTASI AELNSLQDIY DYLEFKYANE 480
481 INEMFINHTG KTGKKHLKNS SYEKFKASPL YQAVSDIGIS AEDVGENISS QHQIHPPVDH 540
541 PSSKPVEVIE SILNANSGDL QVFTSNTKLA IDTVQKYYSL ELSKNTKIRE KVRSDFSKYY 600
601 LADVVLTAKG KKEIQKGSLY EDIKYAINRT PMHFRRDPDV FLKMVEAESL NLLSVKLHMS 660
661 SQAQYIEHLF QIALETTNTS DIAIEWNNFR KLAFNQAMDK IFQDISQEVK DNLTKNCQKL 720
721 VAKTVRHKFM TKLDQAPFIP NVRDPKIPKI LSLTCGQGRF GADAIIAVYV NRKGDFIRDY 780
781 KIVDNPFDKT NPEKFEDTLD NIIQSCQPNA IGINGPNPKT QKFYKRLQEV LHKKQIVDSR 840
841 GHTIPIIYVE DEVAIRYQNS ERAAQEFPNK PPLVKYCIAL ARYMHSPLLE YANLTSEEVR 900
901 SLSIHPHQNL LSSEQLSWAL ETAFVDIVNL VSVEVNKATD NNYYASALKY ISGFGKRKAI 960
961 DFLQSLQRLN EPLLARQQLI THNILHKTIF MNSAGFLYIS WNEKRQKYED LEHDQLDSTR1020
1021 IHPEDYHLAT KVAADALEYD PDTIAEKEEQ GTMSEFIELL REDPDRRAKL ESLNLESYAE1080
1081 ELEKNTGLRK LNNLNTIVLE LLDGFEELRN DFHPLQGDEI FQSLTGESEK TFFKGSIIPV1140
1141 RVERFWHNDI ICTTNSEVEC VVNAQRHAGA QLRRPANEIY EIGKTYPAKV IYIDYANITA1200
1201 EVSLLDHDVK QQYVPISYSK DPSIWDLKQE LEDAEEERKL MMAEARAKRT HRVINHPYYF1260
1261 PFNGRQAEDY LRSKERGEFV IRQSSRGDDH LVITWKLDKD LFQHIDIQEL EKENPLALGK1320
1321 VLIVDNQKYN DLDQIIVEYL QNKVRLLNEM TSSEKFKSGT KKDVVKFIED YSRVNPNKSV1380
1381 YYFSLNHDNP GWFYLMFKIN ANSKLYTWNV KLTNTGYFLV NYNYPSVIQL CNGFKTLLKS1440
1441 NSSKNRMNNY R |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
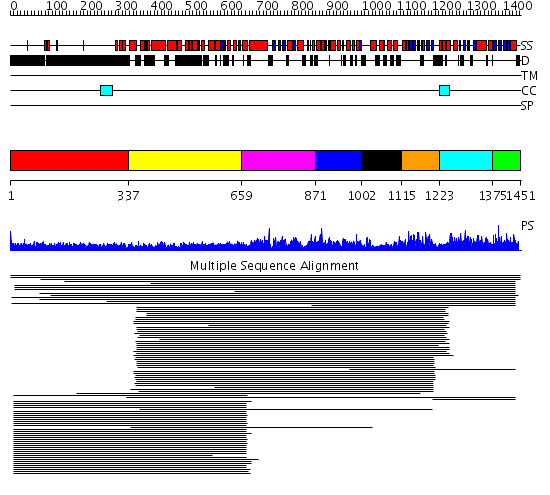
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..336] | 31.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [337..658] | 8.534997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [659..870] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [871..1001] | 1.104997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1002..1114] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1115..1222] | 14.39794 | S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase | |
| 7 | View Details | [1223..1374] | 7.21 | Phosphatidylinositol 3-kinase, p85-alpha subunit | |
| 8 | View Details | [1375..1451] | 1.016979 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)