













| General Information: |
|
| Name(s) found: |
SCC1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 566 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVTENPQRLT VLRLATNKGP LAQIWLASNM SNIPRGSVIQ THIAESAKEI AKASGCDDES 60
61 GDNEYITLRT SGELLQGIVR VYSKQATFLL TDIKDTLTKI SMLFKTSQKM TSTVNRLNTV 120
121 TRVHQLMLED AVTEREVLVT PGLEFLDDTT IPVGLMAQEN SMERKVQGAA PWDTSLEVGR 180
181 RFSPDEDFEH NNLSSMNLDF DIEEGPITSK SWEEGTRQSS RNFDTHENYI QDDDFPLDDA 240
241 GTIGWDLGIT EKNDQNNDDD DNSVEQGRRL GESIMSEEPT DFGFDLDIEK EAPAGNIDTI 300
301 TDAMTESQPK QTGTRRNSKL LNTKSIQIDE ETENSESIAS SNTYKEERSN NLLTPQPTNF 360
361 TTKRLWSEIT ESMSYLPDPI LKNFLSYESL KKRKIHNGRE GSIEEPELNV SLNLTDDVIS 420
421 NAGTNDNSFN ELTDNMSDFV PIDAGLNEAP FPEENIIDAK TRNEQTTIQT EKVRPTPGEV 480
481 ASKAIVQMAK ILRKELSEEK EVIFTDVLKS QANTEPENIT KREASRGFFD ILSLATEGCI 540
541 GLSQTEAFGN IKIDAKPALF ERFINA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
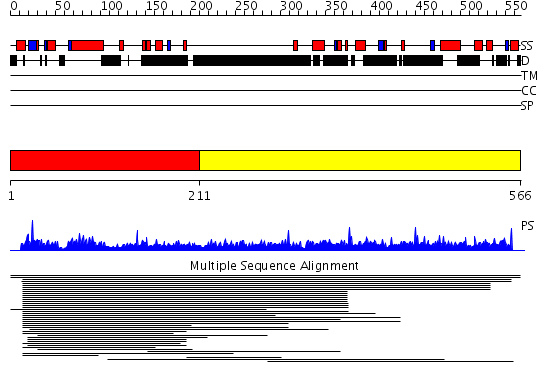
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 5.024974 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [211..566] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [219..380] | 1.070972 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [381..566] | 46.045757 | Sc Smc1hd:Scc1-C complex, ATPgS |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)