













| General Information: |
|
| Name(s) found: |
RAD57_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 460 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPRALSIKFD NTYMDLYDEL PESKLLYDEE FSYLLDAVRQ NGVCVVDFLT LTPKELARLI 60
61 QRSINEVFRF QQLLVHEYNE KYLEICEKNS ISPDNGPECF TTADVAMDEL LGGGIFTHGI 120
121 TEIFGESSTG KSQLLMQLAL SVQLSEPAGG LGGKCVYITT EGDLPTQRLE SMLSSRPAYE 180
181 KLGITQSNIF TVSCNDLINQ EHIINVQLPI LLERSKGSIK LVIIDSISHH LRVELQNKSF 240
241 RESQENKNYL DRMAEKLQIL AHDYSLSVVV ANQVGDKPLA NSPVAHRTYV TDYDYQLGWL 300
301 VGWKNSTILY RQMNSLLGAS SNNDEILSDD EDYMLIERVM STVNDRNYDF FSKKKPPIIE 360
361 NKTVERNSSS PISRQSKKRK FDYRVPNLGL TWSNHVSTRI LLQKSFKAST IIQRGEAHLY 420
421 KGGDSASFWQ VKRTMKVVYS TFAKPGQIAY QITKRGIETA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
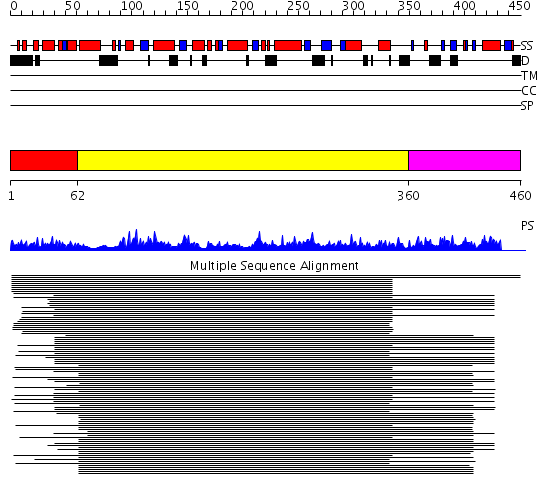
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] | 18.522879 | DNA repair protein Rad51, N-terminal domain | |
| 2 | View Details | [62..359] | 102.67837 | RecA protein, ATPase-domain; RecA protein, C-terminal domain | |
| 3 | View Details | [360..460] | 102.67837 | RecA protein, ATPase-domain; RecA protein, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)