













| General Information: |
|
| Name(s) found: |
KA122_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1081 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSIHEVVAL IEELYSPHPK HDVNQIQQSL QSIQKSEQGF HLANELLSDD KYSANVKYFG 60
61 ALTLTVQLNT RGENDYETLW NVFRSNLLYL TKFSTLYVSN PNMYGQSLII IKKLMSNLSL 120
121 IFTKINDPQL NNAGNENMIK QWNNPINTFI QLMSVQNQNI NADQLLLDSI NCSLTYEQLS 180
181 QFVSLSQKHN ELALTFTEVI VEDLTKFQTK RHSMSQIHEV VHEHLYISTM ALINLNLTAQ 240
241 AVFNPTVFDC ITAWINYISL TRSVSSSGRM DLSEIFQNLI DLMYQSTEGS DGYENAEKIL 300
301 TIFGNVFAND PLLMSYDLRQ QIECIFLGVV RPDSGITDIS NKNSWMLQYM NYLVTNDFFS 360
361 ELKELAICIV DFLQINTLSV CNKLFTNIQA ADNGQVQDEY IQEYIKVLLQ MTNFPLTPVL 420
421 QEFFSVRMVD FWLDLSDAYT NLASETLRPN SIELSTQIFQ QLINIYLPKI SLSVKQRIIE 480
481 EEGESTSVNE FEDFRNAVSD LAQSLWSILG NDNLTNVLID GMGQMPAASD ETLIIKDTDV 540
541 LFRIETMCFV LNTILVDMTL SESPWIKNIV DANKFFNQNV ISVFQTGFQT SASTKVSQIL 600
601 KLDFVRTSTT LIGTLAGYFK QEPFQLNPYV EALFQGLHTC TNFTSKNEQE KISNDKLEVM 660
661 VIKTVSTLCE TCREELTPYL MHFISFLNTV IMPDSNVSHF TRTKLVRSIG YVVQCQVSNG 720
721 PEEQAKYILQ LTNLLSGSIE HCLASSVQLQ EQQDYINCLL YCISELATSL IQPTEIIEND 780
781 ALLQRLSEFQ SFWSSDPLQI RSKIMCTIDK VLDNSIYCKN SAFVEIGCLI VGKGLNLPDG 840
841 EPYFLKYNMS EVMNFVLRHV PNCELATCLP YFVYLLEKLI SEFRKELTPQ EFDFMFEKIL 900
901 LVYYDAYIIN DPDLLQMTIG FVNNVLDVKP GLAIGSKHWT SFILPQFLKL IPSREKFTIV 960
961 AVAKFWTKLI NNKKYNQEEL TTVRQQVSSI GGDLVYQIMY GLFHTQRSDL NSYTDLLRAL1020
1021 VAKFPIEARE WLVAVLPQIC NNPAGHEKFI NKLLITRGSR AAGNVILQWW LDCTTLPNYQ1080
1081 G |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
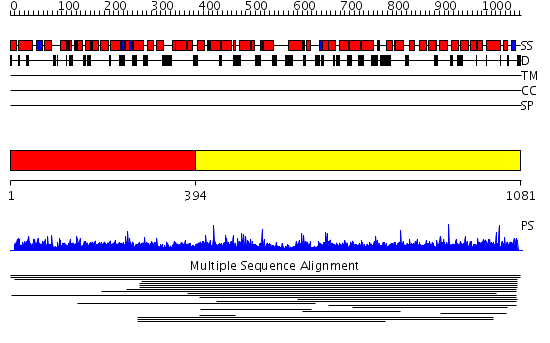
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..393] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [394..1081] | 2.014875 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [929..1081] | 2.079965 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)