













| General Information: |
|
| Name(s) found: |
IMB2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 918 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASTWKPAED YVLQLATLLQ NCMSPNPEIR NNAMEAMENF QLQPEFLNYL CYILIEGESD 60
61 DVLKQHYSLQ DLQNNRATAG MLLKNSMLGG NNLIKSNSHD LGYVKSNIIH GLYNSNNNLV 120
121 SNVTGIVITT LFSTYYRQHR DDPTGLQMLY QLLELTSNGN EPSIKALSKI MEDSAQFFQL 180
181 EWSGNTKPME ALLDSFFRFI SNPNFSPVIR SESVKCINTV IPLQTQSFIV RLDKFLEIIF 240
241 QLAQNDENDL VRAQICISFS FLLEFRPDKL VSHLDGIVQF MLHLITTVNE EKVAIEACEF 300
301 LHAFATSPNI PEHILQPYVK DIVPILLSKM VYNEESIVLL EASNDDDAFL EDKDEDIKPI 360
361 APRIVKKKEA GNGEDADDNE DDDDDDDDED GDVDTQWNLR KCSAATLDVM TNILPHQVMD 420
421 IAFPFLREHL GSDRWFIREA TILALGAMAE GGMKYFNDGL PALIPFLVEQ LNDKWAPVRK 480
481 MTCWTLSRFS PWILQDHTEF LIPVLEPIIN TLMDKKKDVQ EAAISSVAVF IENADSELVE 540
541 TLFYSQLLTS FDKCLKYYKK KNLIILYDAI GRFAEKCALD ETAMQIILPP LIEKWALLSD 600
601 SDKELWPLLE CLSCVASSLG ERFMPMAPEV YNRAFRILCH CVELEAKSHQ DPTIVVPEKD 660
661 FIITSLDLID GLVQGLGAHS QDLLFPQGTK DLTILKIMLE CLQDPVHEVR QSCFALLGDI 720
721 VYFFNSELVI GNLEDFLKLI GTEIMHNDDS DGTPAVINAI WALGLISERI DLNTYIIDMS 780
781 RIILDLFTTN TQIVDSSVME NLSVTIGKMG LTHPEVFSSG AFANDSNWNK WCLSVNALDD 840
841 VEEKSSAYMG FLKIINLTST EVTMSNDTIH KIVTGLSSNV EANVFAQEIY TFLMNHSAQI 900
901 SAINFTPDEI SFLQQFTS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
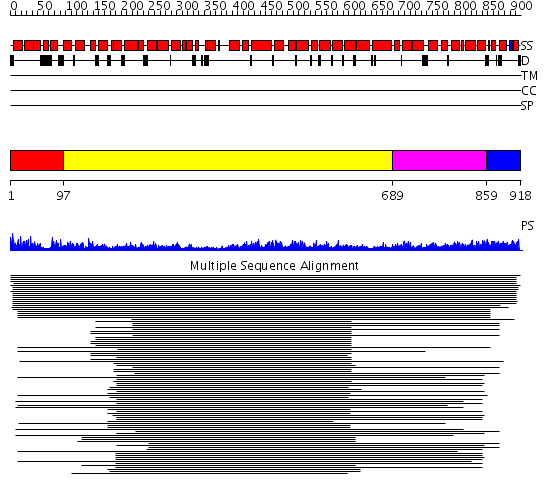
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [97..688] | 20.431997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [689..858] | 2.262988 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [859..918] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)