













| General Information: |
|
| Name(s) found: |
ELG1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 791 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRHVSLSDI LTGNKRKVRR QDALQITIDD ENDTESGTFD ARTAKHDDSS VIFLNHSVVK 60
61 PIEAVSTNHK SAKEFLMTKR TKEKCDDDDD DLIVISDKSP KSETNCSKIA LSQEHEDDIS 120
121 IISTSRIKSS LLNERASKIK NFLKHETTDT FKRLNSISKL NEIEPPLPLH QSIFPVGDKE 180
181 LSDRSVDIPL PFRTIPPLNH NFLPSDYESL KDKNSASCIP VRYQAPVLLG TNIKRNTTLT 240
241 WPQLFKPVTL KQVLIEPKLK LRIKNWIETS FHTLEKPTLR NRLLNRINPN KQQGSGDELA 300
301 NFIVPDLEED ENLRPDFYRN GEANSSLSEF VPLMILHGNS IGKKTLIQTI MREIAGDDNS 360
361 YQIYEVNSNM NRSKKDLLDI LLDFTTTHYV KDSSKRKSDY GLVLFNDVDV LFKEHDRGYW 420
421 AMISKLCEFS RRPLVLTCKD LSLVPSELIA LASEQNSLFH TKKISTSTVY AFLTKYLKSL 480
481 EIEVCDDWLR DVVKQNNADI RKCLMHLQFW CVDTEADLIS SKNRLPVLTS TLGSSVKDIS 540
541 QLTDLLSIND VIGQATLNRS MVRQEIDSTT MTPEKVNTFQ DQNLDDEMKL KFDYVIDYKL 600
601 HLNDPNRQPL LPFELNIYQH IQEQLEARYS YVREANHRLD NEYLVNRFKK MTESTLNFLA 660
661 SRIPKYDHLQ SARRTRNSKK ISDILNQFKG IYNDETLNEN AEIDLLSATT QQIKAEINPF 720
721 VFEIAKSDAN VKNENKQIFE LHSENVSERR YKDLVYQLSQ EGVLKNVWFN ADPSIVVRKW 780
781 EHLHSGFSKN K |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
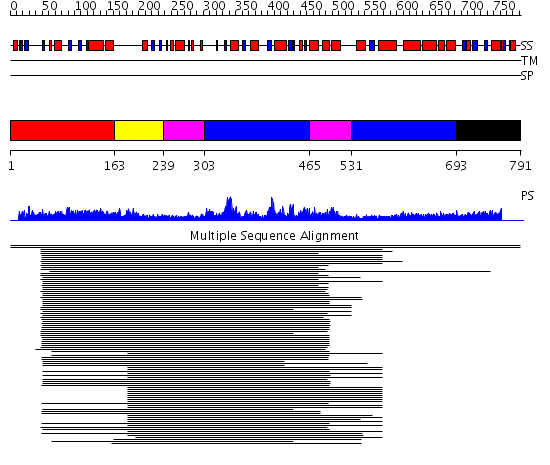
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | 30.0 | ClpB | |
| 2 | View Details | [163..238] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [239..302] [465..530] |
68.0 | Replication factor C | |
| 4 | View Details | [303..464] [531..692] |
68.0 | Replication factor C | |
| 5 | View Details | [693..791] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)