













| General Information: |
|
| Name(s) found: |
COPB_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 973 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSLSSQPAY TLVFDPSPSM ETYSSTDFQK ALEKGSDEQK IDTMKSILVT MLEGNPMPEL 60
61 LMHIIRFVMP SKNKELKKLL YFYWEIVPKL AEDGKLRHEM ILVCNAIQHD LQHPNEYIRG 120
121 NTLRFLTKLR EAELLEQMVP SVLACLEYRH AYVRKYAILA VFSIFKVSEH LLPDAKEIIN 180
181 SFIVAETDPI CKRNAFIGLA ELDRENALHY LENNIADIEN LDPLLQAVFV QFIRQDANRT 240
241 PALKAQYIEL LMELLSTTTS DEVIFETALA LTVLSANPNV LVPAVNKLID LAVKVSDNNI 300
301 KLIVLDRIQD INANNVGALE ELTLDILRVL NAEDLDVRSK ALDISMDLAT SRNAEDVVQL 360
361 LKKELQTTVN NPDQDKAMQY RQLLIKTIRT VAVNFVEMAA SVVSLLLDFI GDLNSVAASG 420
421 IIAFIKEVIE KYPQLRANIL ENMVQTLDKV RSAKAYRGAL WIMGEYAEGE SEIQHCWKHI 480
481 RNSVGEVPIL QSEIKKLTQN QEHTEENEVD ATAKPTGPVI LPDGTYATES AFDVKTSQKS 540
541 VTDEERDSRP PIRRFVLSGD FYTAAILANT IIKLVLKFEN VSKNKTVINA LKAEALLILV 600
601 SIVRVGQSSL VEKKIDEDSL ERVMTSISIL LDEVNPEEKK EEVKLLEVAF LDTTKSSFKR 660
661 QIEIAKKNKH KRALKDSCKN IEPIDTPISF RQFAGVDSTN VQKDSIEEDL QLAMKGDAIH 720
721 ATSSSSISKL KKIVPLCGFS DPVYAEACIT NNQFDVVLDV LLVNQTKETL KNLHVQFATL 780
781 GDLKIIDTPQ KTNVIPHGFH KFTVTVKVSS ADTGVIFGNI IYDGAHGEDA RYVILNDVHV 840
841 DIMDYIKPAT ADDEHFRTMW NAFEWENKIS VKSQLPTLHA YLRELVKGTN MGILTPSESL 900
901 GEDDCRFLSC NLYAKSSFGE DALANLCIEK DSKTNDVIGY VRIRSKGQGL ALSLGDRVAL 960
961 IAKKTNKLAL THV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
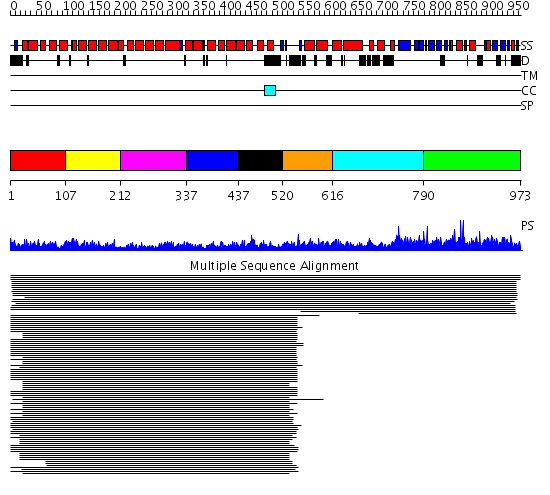
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 266.0 | Adaptin beta subunit N-terminal fragment | |
| 2 | View Details | [107..211] | 266.0 | Adaptin beta subunit N-terminal fragment | |
| 3 | View Details | [212..336] | 266.0 | Adaptin beta subunit N-terminal fragment | |
| 4 | View Details | [337..436] | 266.0 | Adaptin beta subunit N-terminal fragment | |
| 5 | View Details | [437..519] | 266.0 | Adaptin beta subunit N-terminal fragment | |
| 6 | View Details | [520..615] | 29.154902 | beta-Catenin | |
| 7 | View Details | [616..789] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [790..973] | 1.01295 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)