













| General Information: |
|
| Name(s) found: |
CDC13_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 924 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTLEEPECP PHKNRIFVSS SKDFEGYPSK AIVPVQFVAL LTSIHLTETK CLLGFSNFER 60
61 RGDQSQEDQY LIKLKFKDRG SERLARITIS LLCQYFDIEL PDLDSDSGAS PTVILRDIHL 120
121 ERLCFSSCKA LYVSKHGNYT LFLEDIKPLD LVSVISTIST KSTNSSKHSS SELISECDLN 180
181 NSLVDIFNNL IEMNRDEKNR FKFVKLIHYD IELKKFVQDQ QKVLSQKSKA AAINPFFVPN 240
241 RLGIPYIESQ NEFNSQLMTL NVDEPTTDIS NMGEEMHDSA DPIEDSDSST TSSTGKYFSS 300
301 KSYIQSQTPE RKTSVPNNWH DDDSGSKRKR KLSFHSPNAS SIRKAISYEQ LSLASVGSVE 360
361 RLEGKIVGMN PPQFASINEF KYCTLKLYFT QLLPNVPDKV LVPGVNCIEI VIPTRERICE 420
421 LFGVLNCQSD KISDILLLEK PDRISVEVER ILWDNDKTAS PGMAVWSLKN ISTDTQAQAQ 480
481 VQVPAQSSAS IDPSRTRMSK MARKDPTIEF CQLGLDTFET KYITMFGMLV SCSFDKPAFI 540
541 SFVFSDFTKN DIVQNYLYDR YLIDYENKLE LNEGFKAIMY KNQFETFDSK LRKIFNNGLR 600
601 DLQNGRDENL SQYGIVCKMN IKVKMYNGKL NAIVRECEPV PHSQISSIAS PSQCEHLRLF 660
661 YQRAFKRIGE SAISRYFEEY RRFFPIHRNG SHLAKLRFDE VKHEPKKSPT TPALAEHIPD 720
721 LNADVSSFDV KFTDISSLLD SSARLPRPQQ THKSNTLYSC EGRIIAIEYH ASDLCFHITN 780
781 ELPLLQTRGL APERVLQLHI ITSKNFAYFF NRSSAYLQRQ PLEEKYTQLA QFLGHSFKFN 840
841 ITSSLTLFPD TTVALQIWCP IECTFRELQQ QLAHPKVAAA PDSGSLDCAI NATVNPLRLL 900
901 AAQNGVTVKK EEDNDDDAGA VPTS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
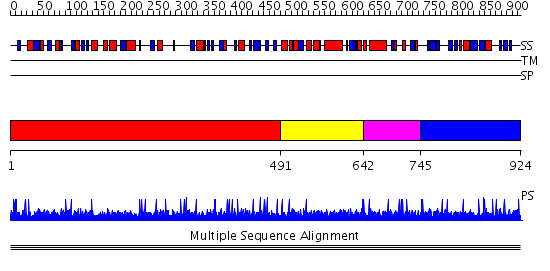
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..490] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [491..641] | 1276.0 | CDC13 ssDNA-binding domain | |
| 3 | View Details | [642..744] | 1276.0 | CDC13 ssDNA-binding domain | |
| 4 | View Details | [745..924] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [706..924] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)