













| General Information: |
|
| Name(s) found: |
ACS1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 713 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPSAVQSSK LEEQSSEIDK LKAKMSQSAA TAQQKKEHEY EHLTSVKIVP QRPISDRLQP 60
61 AIATHYSPHL DGLQDYQRLH KESIEDPAKF FGSKATQFLN WSKPFDKVFI PDPKTGRPSF 120
121 QNNAWFLNGQ LNACYNCVDR HALKTPNKKA IIFEGDEPGQ GYSITYKELL EEVCQVAQVL 180
181 TYSMGVRKGD TVAVYMPMVP EAIITLLAIS RIGAIHSVVF AGFSSNSLRD RINDGDSKVV 240
241 ITTDESNRGG KVIETKRIVD DALRETPGVR HVLVYRKTNN PSVAFHAPRD LDWATEKKKY 300
301 KTYYPCTPVD SEDPLFLLYT SGSTGAPKGV QHSTAGYLLG ALLTMRYTFD THQEDVFFTA 360
361 GDIGWITGHT YVVYGPLLYG CATLVFEGTP AYPNYSRYWD IIDEHKVTQF YVAPTALRLL 420
421 KRAGDSYIEN HSLKSLRCLG SVGEPIAAEV WEWYSEKIGK NEIPIVDTYW QTESGSHLVT 480
481 PLAGGVTPMK PGSASFPFFG IDAVVLDPNT GEELNTSHAE GVLAVKAAWP SFARTIWKNH 540
541 DRYLDTYLNP YPGYYFTGDG AAKDKDGYIW ILGRVDDVVN VSGHRLSTAE IEAAIIEDPI 600
601 VAECAVVGFN DDLTGQAVAA FVVLKNKSSW STATDDELQD IKKHLVFTVR KDIGPFAAPK 660
661 LIILVDDLPK TRSGKIMRRI LRKILAGESD QLGDVSTLSN PGIVRHLIDS VKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
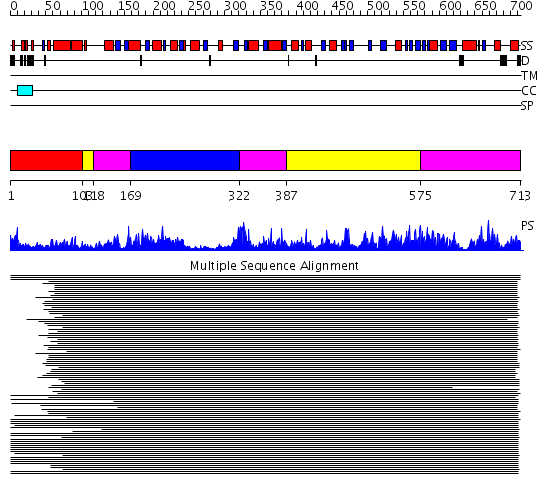
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 3.509997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [103..117] [387..574] |
277.218487 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 3 | View Details | [118..168] [322..386] [575..713] |
277.218487 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 4 | View Details | [169..321] | 277.218487 | Phenylalanine activating domain of gramicidin synthetase 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)