













| General Information: |
|
| Name(s) found: |
dnaJ /
EG10240
[EcoGene]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli str. K-12 substr. MG1655 |
| Length: | 376 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane fraction
[IDA cytoplasm [IDA |
| Biological Process: |
response to heat
[IEA][IEP DNA replication [IEA][IMP protein refolding [IDA protein folding [IDA response to stress [IEA] |
| Molecular Function: |
heat shock protein binding
[IEA]
ATP binding [IEA] zinc ion binding [IDA protein homodimerization activity [IDA protein binding [IMP protein disulfide isomerase activity [IDA unfolded protein binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKQDYYEIL GVSKTAEERE IRKAYKRLAM KYHPDRNQGD KEAEAKFKEI KEAYEVLTDS 60
61 QKRAAYDQYG HAAFEQGGMG GGGFGGGADF SDIFGDVFGD IFGGGRGRQR AARGADLRYN 120
121 MELTLEEAVR GVTKEIRIPT LEECDVCHGS GAKPGTQPQT CPTCHGSGQV QMRQGFFAVQ 180
181 QTCPHCQGRG TLIKDPCNKC HGHGRVERSK TLSVKIPAGV DTGDRIRLAG EGEAGEHGAP 240
241 AGDLYVQVQV KQHPIFEREG NNLYCEVPIN FAMAALGGEI EVPTLDGRVK LKVPGETQTG 300
301 KLFRMRGKGV KSVRGGAQGD LLCRVVVETP VGLNERQKQL LQELQESFGG PTGEHNSPRS 360
361 KSFFDGVKKF FDDLTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
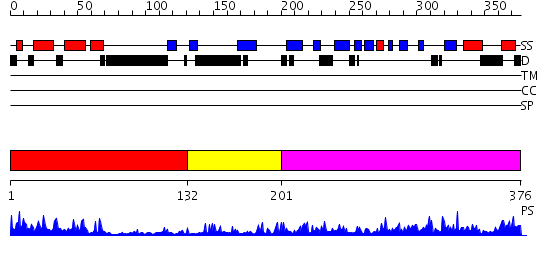
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 23.0 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [132..200] | 15.39794 | Cysteine-rich domain of the chaperone protein DnaJ. | |
| 3 | View Details | [201..376] | 6.27 | No description for 2b26A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)