













| General Information: |
|
| Name(s) found: |
NDUS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 748 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrial respiratory chain complex I [IDA] mitochondrial membrane [IDA] respiratory chain complex I [IDA] mitochondrion [IDA] |
| Biological Process: |
response to oxidative stress
[IDA]
embryonic development ending in seed dormancy [NAS] photorespiration [TAS] |
| Molecular Function: |
NADH dehydrogenase activity
[ISS]
oxidoreductase activity [IEA] electron carrier activity [IEA] NADH dehydrogenase (ubiquinone) activity [IEA] oxidoreductase activity, acting on NADH or NADPH [IEA] iron-sulfur cluster binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLGILASRT IRPASRLLQS QTSNFFLRTI VSKPELQSPE SAAVSEPEPP TQILPPRNPV 60
61 GGARVHFSNP EDAIEVFVDG YAVKVPKGFT VLQACEVAGV DIPRFCYHSR LSIAGNCRMC 120
121 LVEVEKSPKP VASCAMPALP GMKIKTDTPI AKKAREGVME FLLMNHPLDC PICDQGGECD 180
181 LQDQSMAFGS DRGRFTEMKR SVVDKNLGPL VKTVMTRCIQ CTRCVRFASE VAGVQDLGIL 240
241 GRGSGEEIGT YVEKLMTSEL SGNVIDICPV GALTSKPFAF KARNWELKAT ETIDVSDAVG 300
301 SNIRVDSRGP EVMRIIPRLN EDINEEWISD KTRFCYDGLK RQRLSDPMIR DSDGRFKAVS 360
361 WRDALAVVGD IIHQVKPDEI VGVAGQLSDA ESMMVLKDFV NRMGSDNVWC EGTAAGVDAD 420
421 LRYSYLMNTS ISGLENADLF LLIGTQPRVE AAMVNARICK TVRASNAKVG YVGPPAEFNY 480
481 DCKHLGTGPD TLKEIAEGRH PFCTALKNAK NPAIIVGAGL FNRTDKNAIL SSVESIAQAN 540
541 NVVRPDWNGL NFLLQYAAQA AALDLGLIQQ SAKALESAKF VYLMGADDVN VDKIPKDAFV 600
601 VYQGHHGDKA VYRANVILPA SAFTEKEGTY ENTEGFTQQT VPAVPTVGDA RDDWKIVRAL 660
661 SEVSGVKLPY NSIEGVRSRI KSVAPNLVHT DEREPAAFGP SLKPECKEAM STTPFQTVVE 720
721 NFYMTNSITR ASKIMAQCSA VLLKKPFV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
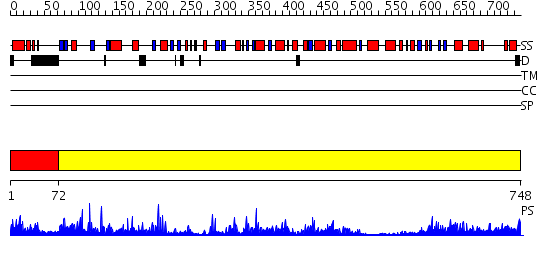
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | 12.30103 | PHTHALATE DIOXYGENASE REDUCTASE: A MODULAR STRUCTURE FOR ELECTRON TRANSFER FROM PYRIDINE NUCLEOTIDES TO [2FE-2S] | |
| 2 | View Details | [72..748] | 94.0 | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)