













| General Information: |
|
| Name(s) found: |
KU70_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 621 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
DNA-dependent protein kinase-DNA ligase 4 complex [IEA] |
| Biological Process: |
DNA repair
[TAS]
response to heat [IEP] telomere maintenance [TAS] |
| Molecular Function: |
protein binding
[IPI]
double-stranded DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELDPDDVFR DEDEDPENDF FQEKEASKEF VVYLIDASPK MFCSTCPSEE EDKQESHFHI 60
61 AVSCIAQSLK AHIINRSNDE IAICFFNTRE KKNLQDLNGV YVFNVPERDS IDRPTARLIK 120
121 EFDLIEESFD KEIGSQTGIV SDSRENSLYS ALWVAQALLR KGSLKTADKR MFLFTNEDDP 180
181 FGSMRISVKE DMTRTTLQRA KDAQDLGISI ELLPLSQPDK QFNITLFYKD LIGLNSDELT 240
241 EFMPSVGQKL EDMKDQLKKR VLAKRIAKRI TFVICDGLSI ELNGYALLRP AIPGSITWLD 300
301 STTNLPVKVE RSYICTDTGA IMQDPIQRIQ PYKNQNIMFT VEELSQVKRI STGHLRLLGF 360
361 KPLSCLKDYH NLKPSTFLYP SDKEVIGSTR AFIALHRSMI QLERFAVAFY GGTTPPRLVA 420
421 LVAQDEIESD GGQVEPPGIN MIYLPYANDI RDIDELHSKP GVAAPRASDD QLKKASALMR 480
481 RLELKDFSVC QFANPALQRH YAILQAIALD ENELRETRDE TLPDEEGMNR PAVVKAIEQF 540
541 KQSIYGDDPD EESDSGAKEK SKKRKAGDAD DGKYDYIELA KTGKLKDLTV VELKTYLTAN 600
601 NLLVSGKKEV LINRILTHIG K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
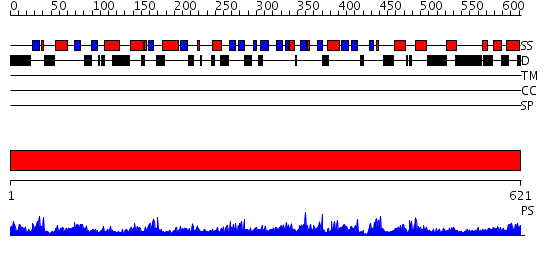
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..621] | 157.0 | DNA binding C-terminal domain of ku70; Ku70 subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)