













| General Information: |
|
| Name(s) found: |
KU80_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 680 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
DNA repair
[TAS]
response to heat [IEP] DNA integration [TAS] telomere maintenance [TAS] double-strand break repair via nonhomologous end joining [TAS] double-strand break repair [IMP] |
| Molecular Function: |
protein binding
[IPI]
double-stranded DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARNREGLVL VLDVGPAMRS VLPDVEKACS MLLQKKLIYN KYDEVGIVVF GTEETGNELA 60
61 REIGGYENVT VLRNIRVVDE LAAEHVKQLP RGTVAGDFLD ALIVGMDMLI KMYGNAHKGK 120
121 KRMCLITNAA CPTKDPFEGT KDDQVSTIAM KMAAEGIKME SIVMRSNLSG DAHERVIEEN 180
181 DHLLTLFSSN AIAKTVNVDS PLSLLGSLKT RRVAPVTLFR GDLEINPTMK IKVWVYKKVA 240
241 EERLPTLKMY SDKAPPTDKF AKHEVKVDYD YKVTAESTEV IAPEERIKGF RYGPQVIPIS 300
301 PDQIETLKFK TDKGMKLLGF TEASNILRHY YMKDVNIVVP DPSKEKSVLA VSAIAREMKE 360
361 TNKVAIVRCV WRNGQGNVVV GVLTPNVSER DDTPDSFYFN VLPFAEDVRE FPFPSFNKLP 420
421 SSWKPDEQQQ AVADNLVKML DLAPSAEEEV LKPDLTPNPV LQRFYEYLEL KSKSTDATLP 480
481 PMDGTFKRLM EQDPELSSNN KSIMDTFRGS FEVKENPKLK KASKRLLRDK PSGSDDEDNR 540
541 MITYDAKENK IDIVGDANPI QDFEAMISRR DKTDWTEKAI TQMKNLIMKL VENCTDEGDK 600
601 ALECVLALRK GCVLEQEPKQ FNEFLNHLFK LCQERNLSHL LEHFMSKKIT LIPKSEAADS 660
661 DIVDENAGDF IVKQESMLES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
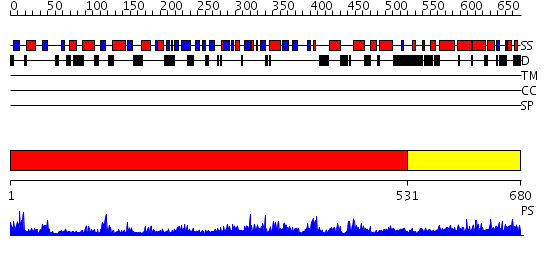
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..530] | 102.0 | Ku80 subunit | |
| 2 | View Details | [531..680] | 45.30103 | Three-dimensional structure of Ku80 CTD |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.62 |
Source: Reynolds et al. (2008)