













| General Information: |
|
| Name(s) found: |
gi|79320679
[NCBI NR]
gi|22655480 [NCBI NR] gi|18408017 [NCBI NR] gi|15983374 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 717 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
protein serine/threonine/tyrosine kinase activity
[ISS]
kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQFRQIGEV LGSLNALMVL QDDILINQRQ CCLLLDIFSL GFNTVAEEIR HNLKLEEKHT 60
61 KWRALEQPLR ELYRVFKEGE MYVRNCMSNK DWWGKVINFH QNKDCVEFHI HNLLCYFSAV 120
121 IEAIETAGEI SGLDPSEMER RRVVFSRKYD REWNDPKLFQ WRFGKQYLVP RDICLRFEHS 180
181 WREDRWNLVE ALQEKRKSKS DEIGKTEKRL ADFLLKKLTG LEQFNGKLFP SSILVGSKDY 240
241 QVRRRLGGGG QYKEIQWLGD SFVLRHFFGD LEPLNAEISS LLSLCHSNIL QYLCGFYDEE 300
301 RKECSLVMEL MHKDLKSYMK ENCGPRRRYL FSVPVVIDIM LQIARGMEYL HSNEIFHGDL 360
361 NPMNILLKER SHTEGYFHAK ISGFGLTSVK NQSFSRASSR PTTPDPVIWY APEVLAEMEQ 420
421 DLKGTAPRSK FTHKADVYSF AMVCFELITG KVPFEDDHLQ GDKMAKNIRT GERPLFPFPS 480
481 PKYLVSLIKR CWHSEPSQRP TFSSICRILR YIKKFLVVNP DHGHLQIQNP LVDCWDLEAR 540
541 FLKKFSMESG SHAGSVTQIP FQLYSYRIAE KEKMSPNFNK EENSDAGESV SESVSVVEEP 600
601 PTTMPMYTKS LCLDAISEYS DTRSVYSEAP IKKTSALKKS GDTIKNRRNS ISGLRSPGSS 660
661 PIKPRSTPKV SSPLSPFGRS SSNKANKDTR LPLSPMSPLS HGRRRQLTGP ASDSELT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
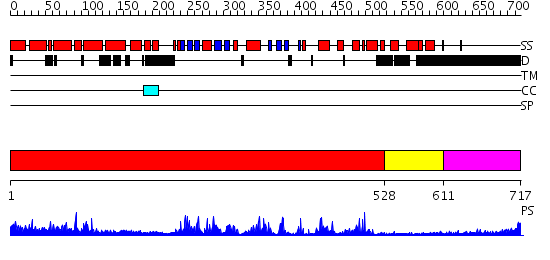
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..527] | 100.0 | CHICKEN SRC TYROSINE KINASE | |
| 2 | View Details | [528..610] | 58.30103 | Crystal Structure of the Auto-Inhibited Kinase Domain of Calcium/Calmodulin Activated Kinase II | |
| 3 | View Details | [611..717] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)